Search Count: 13
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: ZN, NA, GOL, D5M, 3D1, ADE |
 |
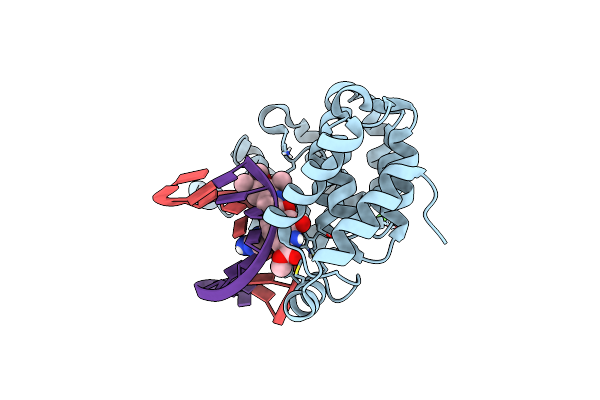
Bacillus Cereus Dna Glycosylase Alkd Bound To A Cc1065-Adenine Nucleobase Adduct And Dna Containing An Abasic Site
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2021-11-17 Classification: HYDROLASE/DNA/ANTIBIOTIC Ligands: CA, YNG |
 |
Bacillus Cereus Dna Glycosylase Alkd Bound To A Duocarmycin Sa-Adenine Nucleobase Adduct And Dna Containing An Abasic Site
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2021-11-17 Classification: HYDROLASE/DNA/ANTIBIOTIC Ligands: CA, YNY |
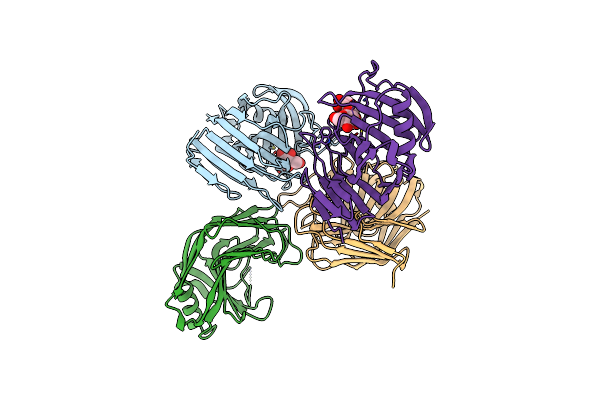 |
Xyloglucanase Domain Of Nopaa, A Type Three Effector From Sinorhizobium Fredii In Complex With Cellobiose
Organism: Sinorhizobium fredii usda 257
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-07-08 Classification: CELL INVASION Ligands: BGC |
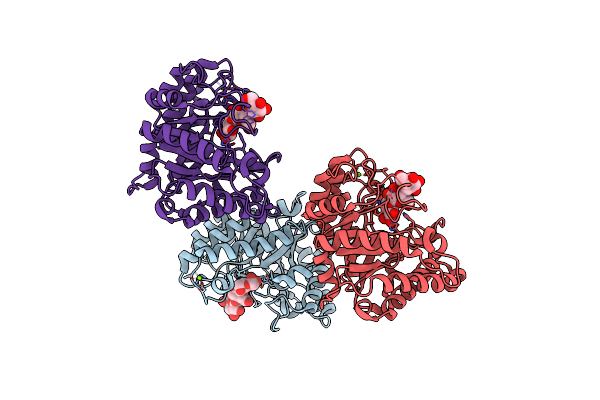 |
Organism: Zobellia galactanivorans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-10-31 Classification: HYDROLASE Ligands: MG |
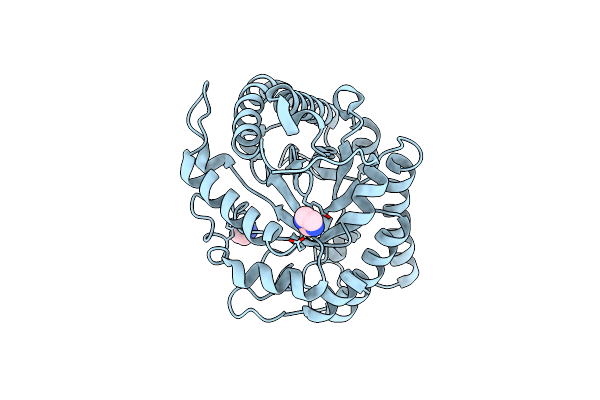 |
Organism: Zobellia galactanivorans
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2018-10-31 Classification: HYDROLASE Ligands: IMD |
 |
Crystal Structure Of A Dual Function Amine Oxidase/Cyclase In Complex With Substrate Analogues
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FAD, ACT, CJ8, CA, GOL, CJE |
 |
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FAD, CA, CWH, GOL, ACT |
 |
Organism: Streptomyces rochei subsp. volubilis
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FAD, CA, CWH, GOL |
 |
Organism: Streptomyces rochei subsp. volubilis
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FAD, OXY, NA |
 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2016-03-23 Classification: PROTEIN BINDING |
 |
1H, 13C, 15N Chemical Shift Assignments Of Streptomyces Virginiae Vira Acp5A
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2014-06-04 Classification: TRANSFERASE |
 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2014-06-04 Classification: RIBOSOMAL PROTEIN |

