Search Count: 85
 |
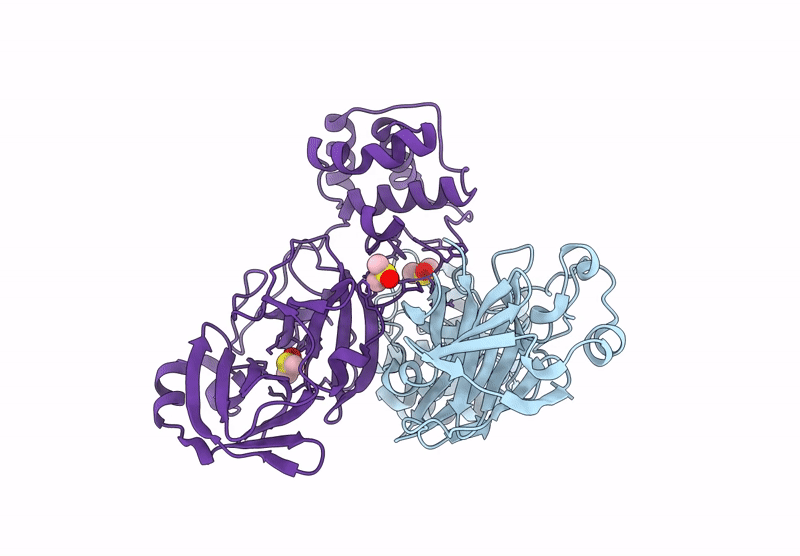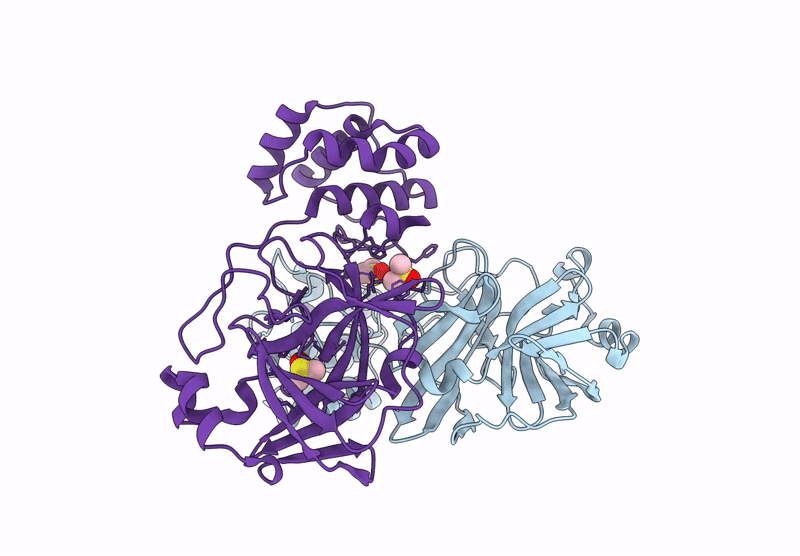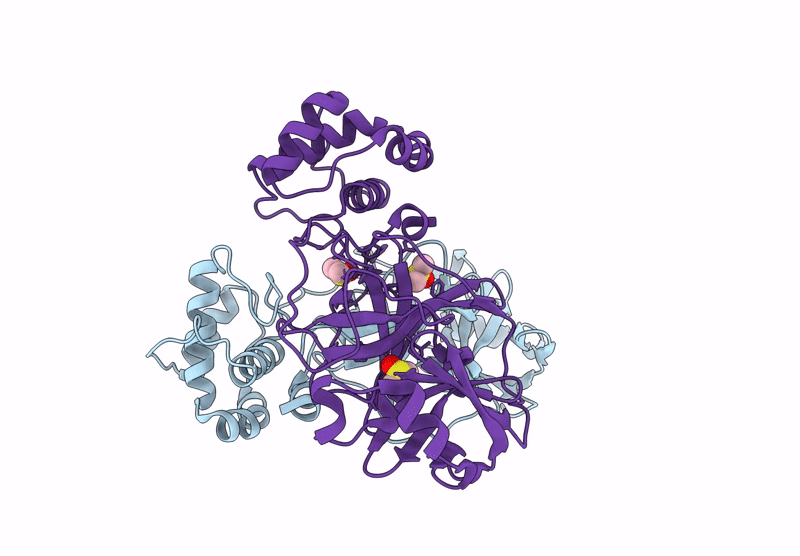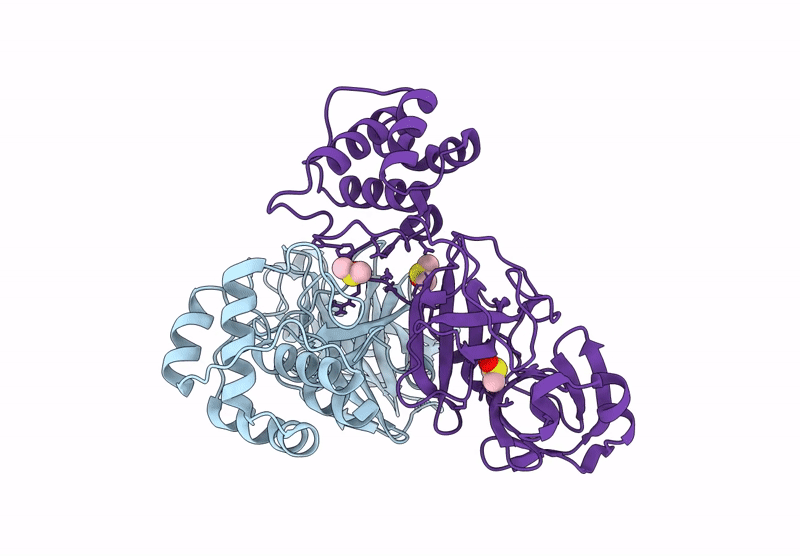
Crystal Structure Of Sars-Cov-2 Main Protease In Orthorhombic Space Group P212121
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-03-22 Classification: HYDROLASE Ligands: CL, DMS, MLI |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-01-25 Classification: VIRAL PROTEIN Ligands: DMS |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-01-18 Classification: VIRAL PROTEIN Ligands: CL |
 |
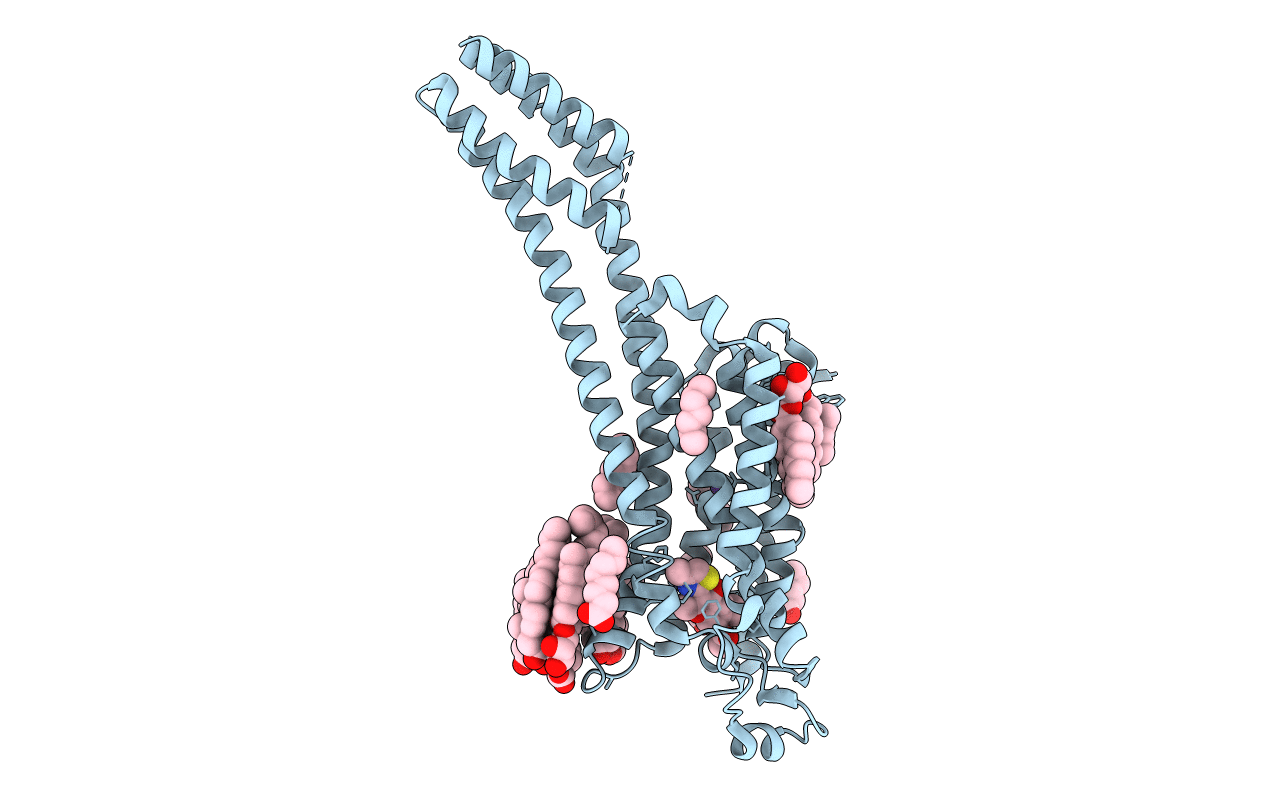
Crystal Structure Of Stabilized A2A Adenosine Receptor A2Ar-Star2-Bril In Complex With Chromone 4D
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2020-09-16 Classification: PROTEIN BINDING Ligands: QGE, NA, CLR, OLA, OLC |
 |
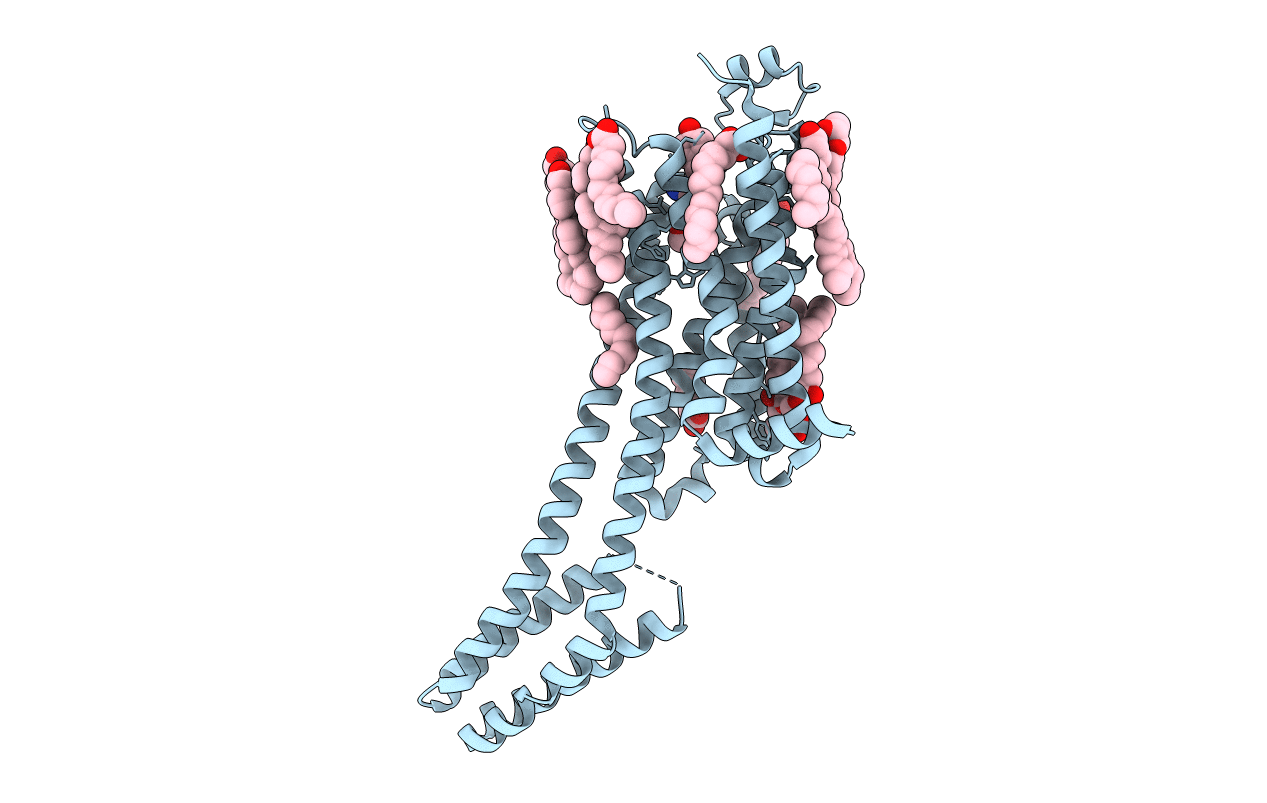
Crystal Structure Of Stabilized A2A Adenosine Receptor A2Ar-Star2-Bril In Complex With Chromone 5D
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2020-09-16 Classification: PROTEIN BINDING Ligands: QGW, NA, CLR, OLB, OLA, OLC |
 |
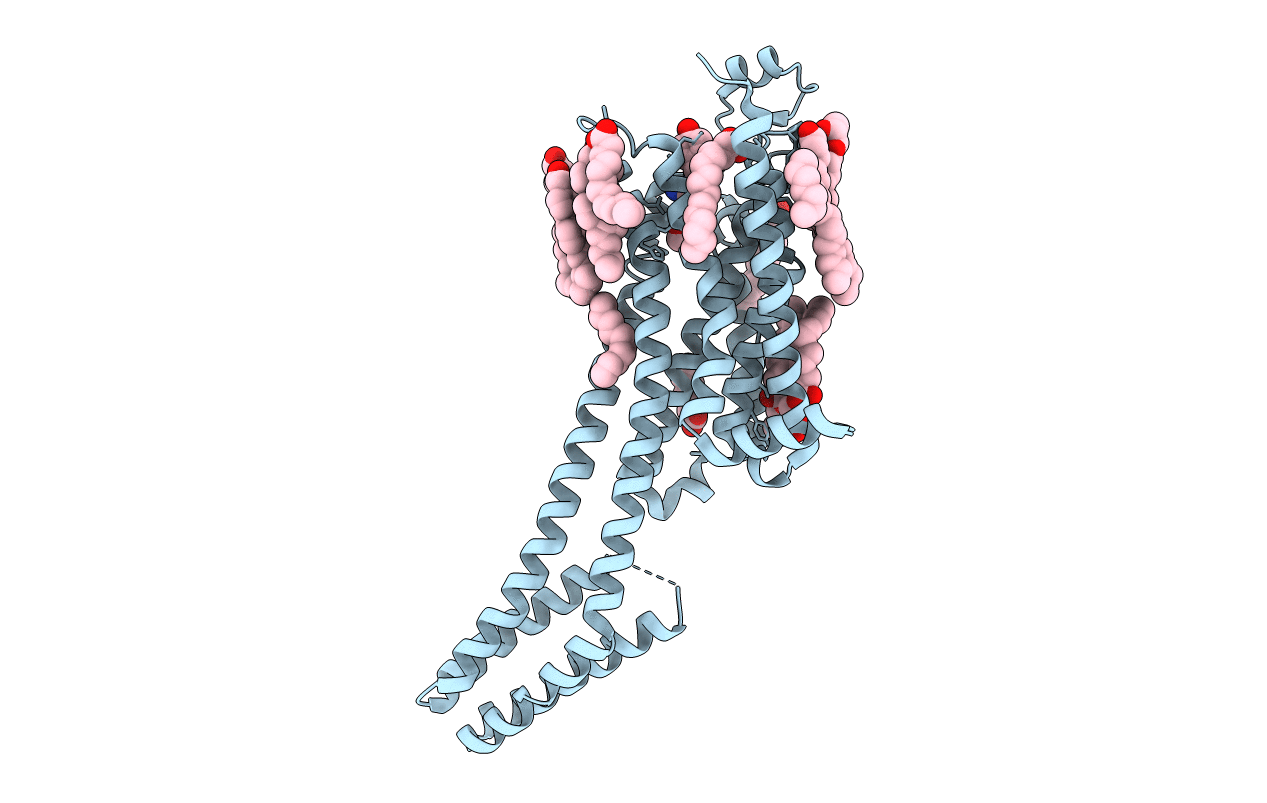
Structure Of The A2A Adenosine Receptor Determined At Swissfel Using Native-Sad At 4.57 Kev From All Available Diffraction Patterns
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2020-07-15 Classification: SIGNALING PROTEIN Ligands: ZMA, OLA, OLB, OLC, CLR |
 |
Structure Of The A2A Adenosine Receptor Determined At Swissfel Using Native-Sad At 4.57 Kev From 50,000 Diffraction Patterns
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2020-07-15 Classification: MEMBRANE PROTEIN Ligands: ZMA, OLA, OLB, OLC, CLR |
 |
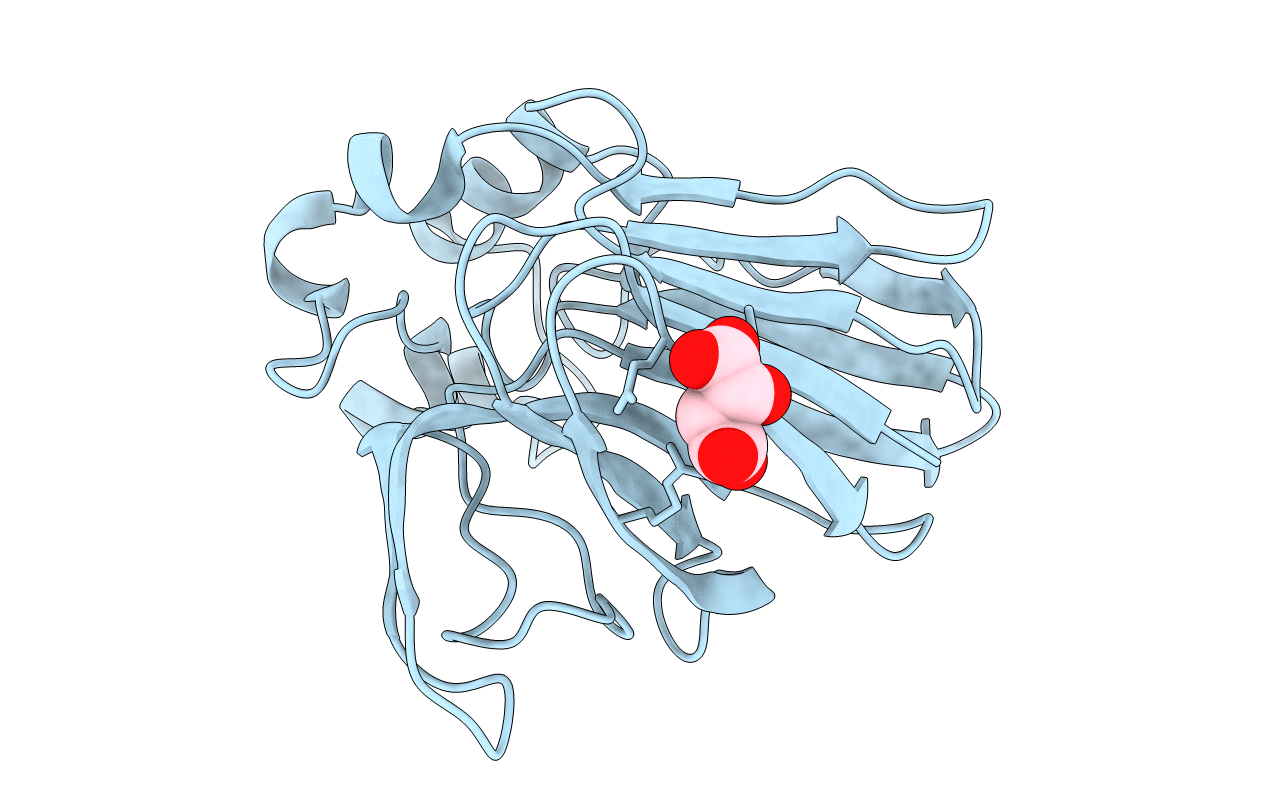
Structure Of Thaumatin Determined At Swissfel Using Native-Sad At 4.57 Kev From All Available Diffraction Patterns
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2020-07-15 Classification: PLANT PROTEIN Ligands: TLA |
 |
Structure Of Thaumatin Determined At Swissfel Using Native-Sad At 4.57 Kev From 20,000 Diffraction Patterns
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2020-07-15 Classification: PLANT PROTEIN Ligands: TLA |
 |
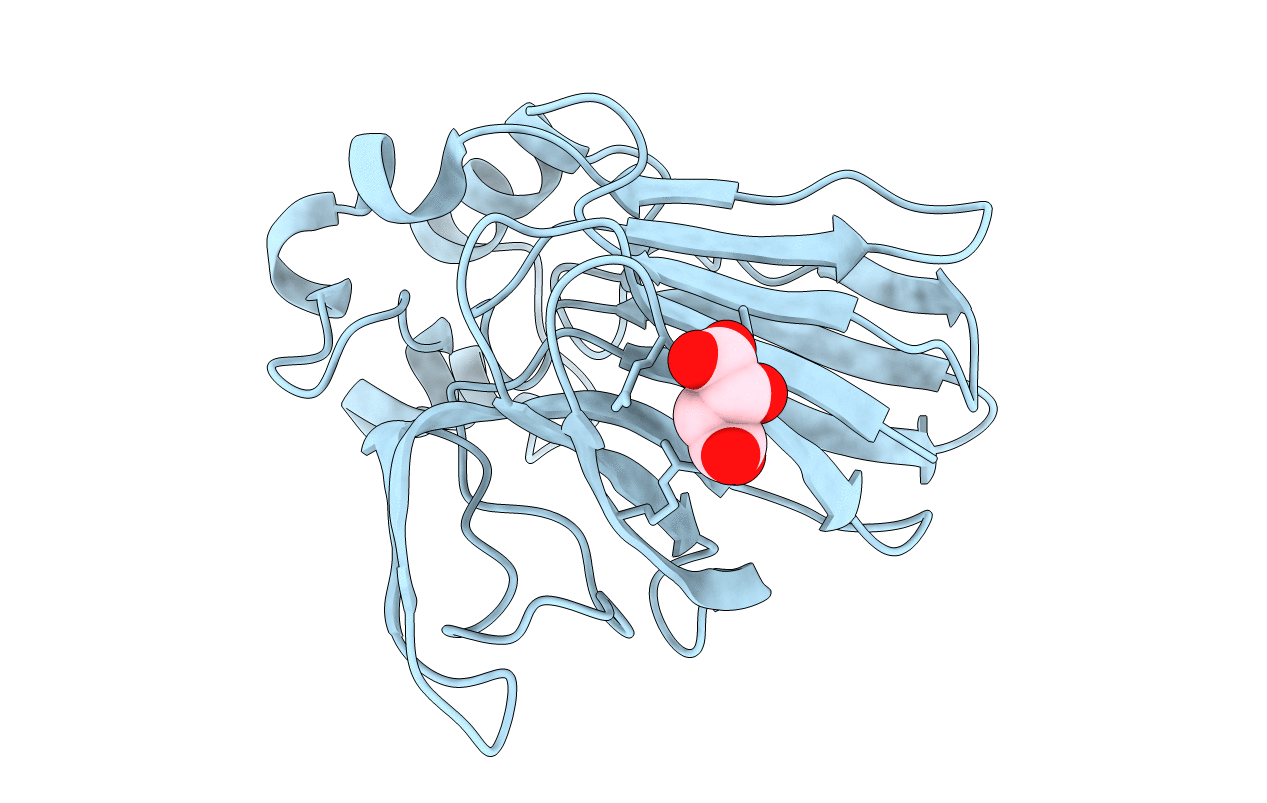
Structure Of Thaumatin Determined At Swissfel Using Native-Sad At 6.06 Kev From All Available Diffraction Patterns
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-07-15 Classification: PLANT PROTEIN Ligands: TLA |
 |
Structure Of Thaumatin Determined At Swissfel Using Native-Sad At 6.06 Kev From 50,000 Diffraction Patterns.
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-15 Classification: PLANT PROTEIN Ligands: TLA |
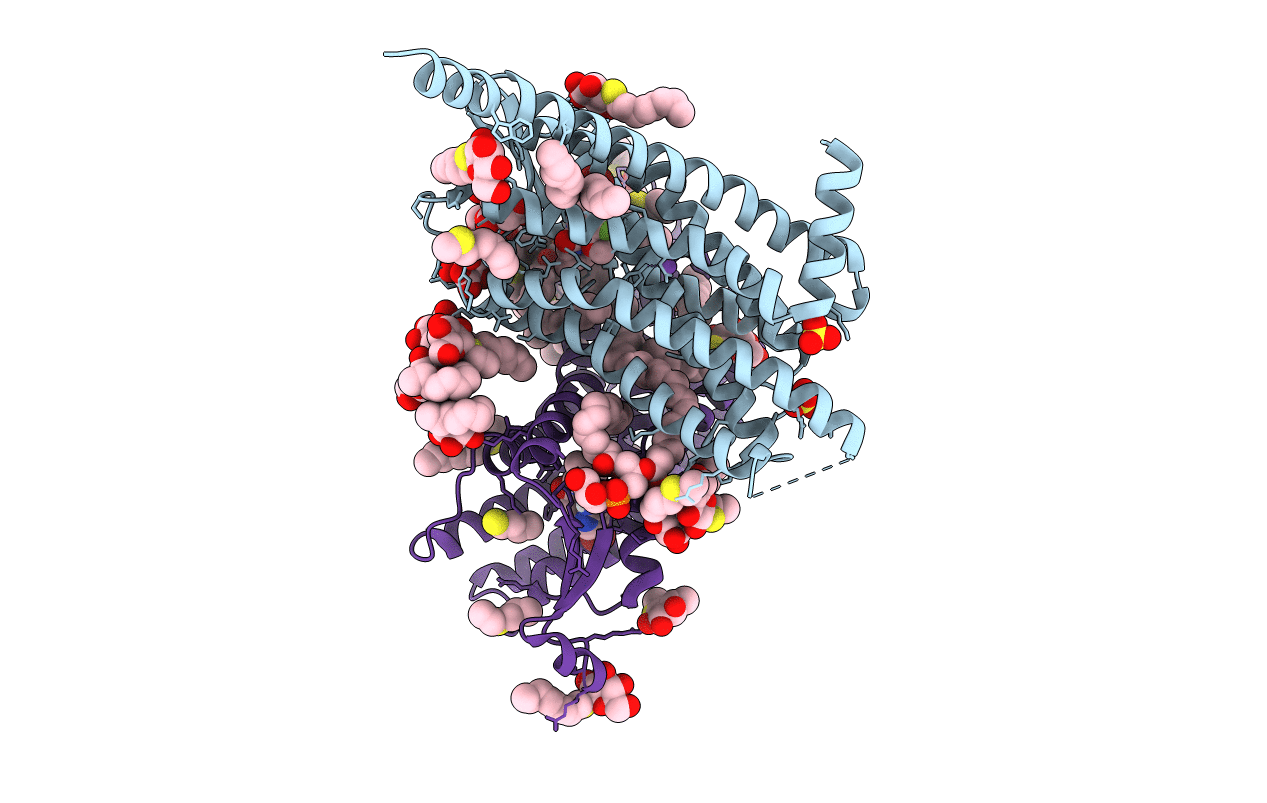 |
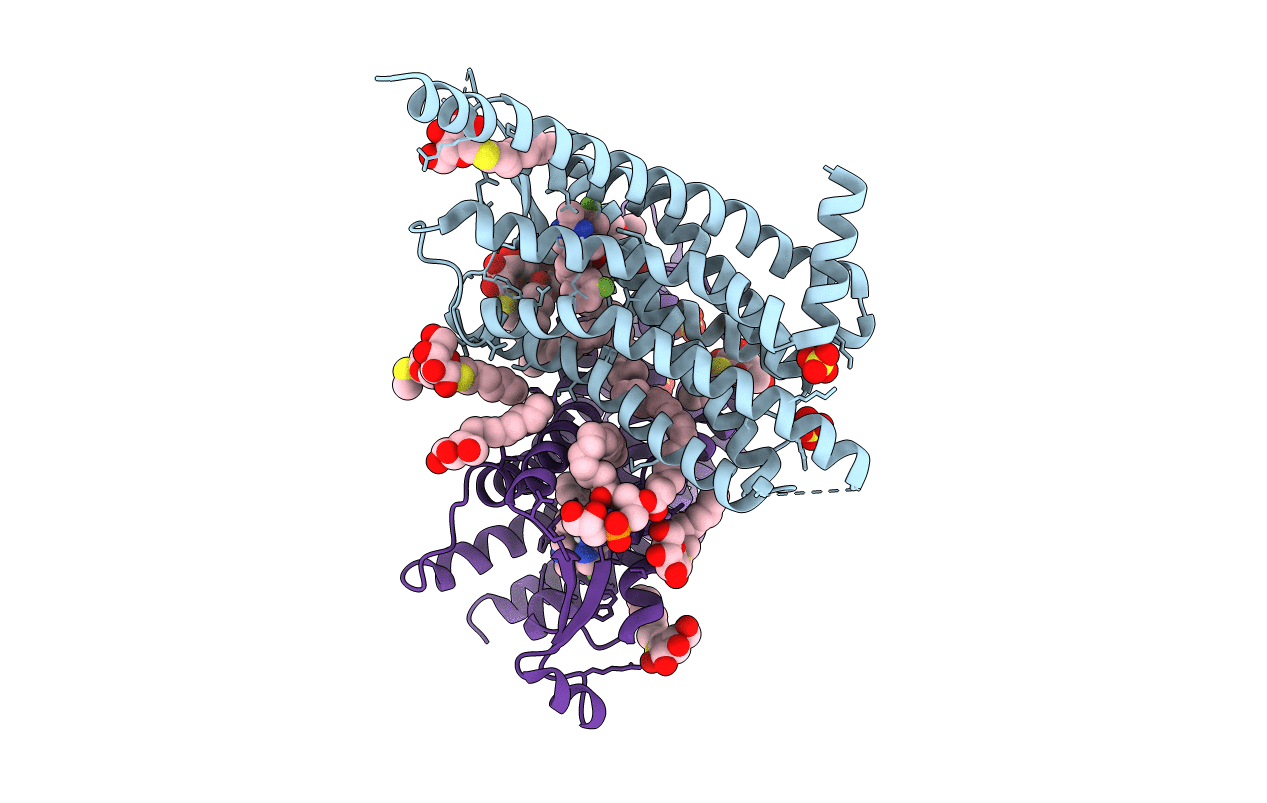
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2020-01-15 Classification: MEMBRANE PROTEIN Ligands: NRE, PG4, CIT, SO4, SOG, NA, PGW |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2020-01-15 Classification: MEMBRANE PROTEIN Ligands: NRK, PG4, SO4, SOG, PGW |
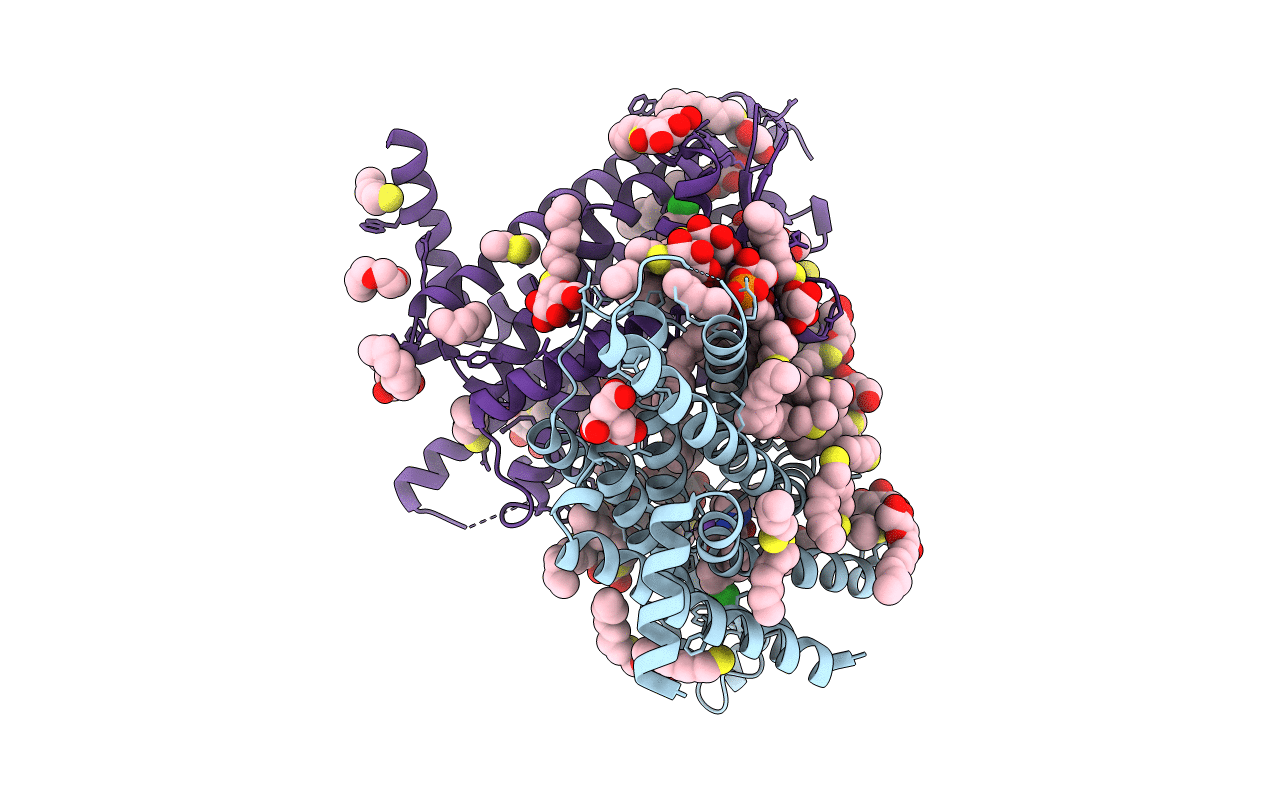 |
Crystal Structure Of The Orexin-1 Receptor In Complex With Suvorexant At 2.29 A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2020-01-01 Classification: MEMBRANE PROTEIN Ligands: SUV, PG4, CIT, SOG, PGW, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2020-01-01 Classification: MEMBRANE PROTEIN Ligands: 7MA, SO4, PG4, CIT, SOG, PGW, NA |
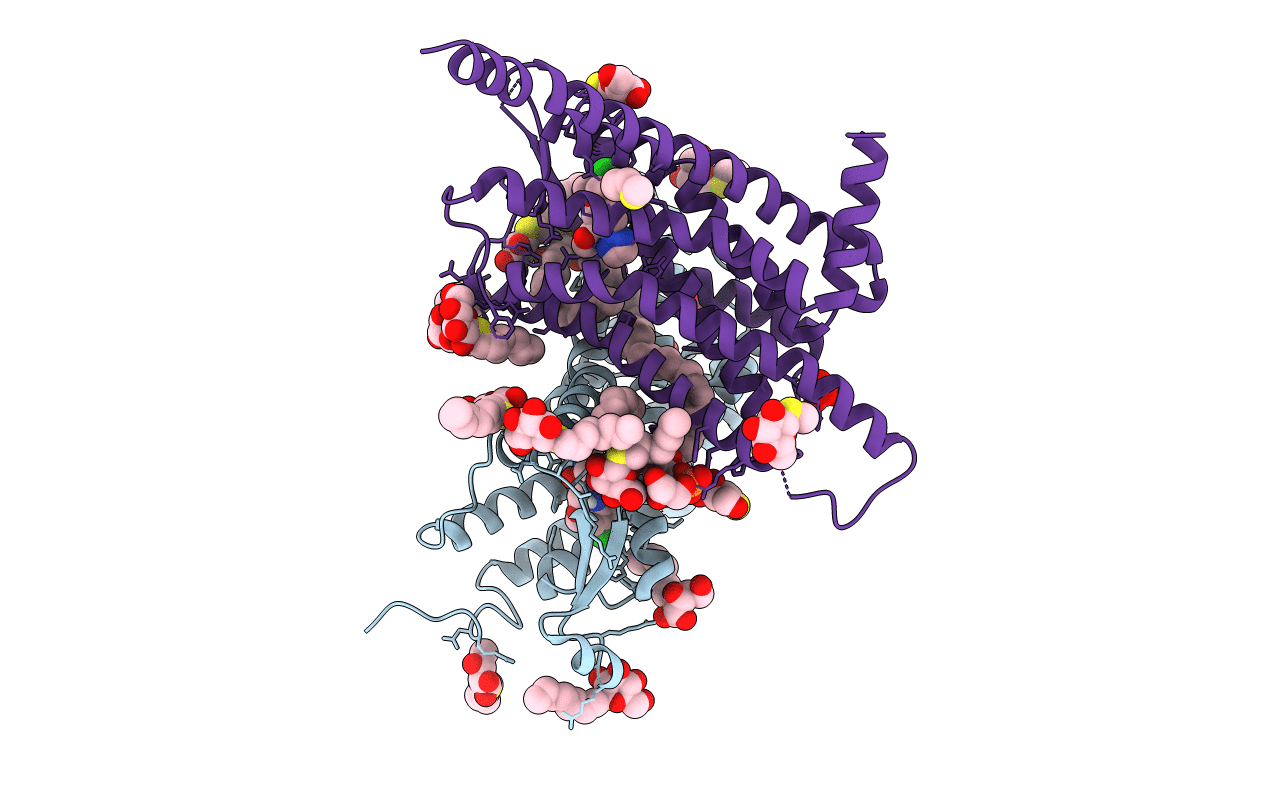 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2020-01-01 Classification: MEMBRANE PROTEIN Ligands: NS2, SO4, PGW, SOG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2020-01-01 Classification: MEMBRANE PROTEIN Ligands: NRZ, SOG, SO4, PGW |
 |
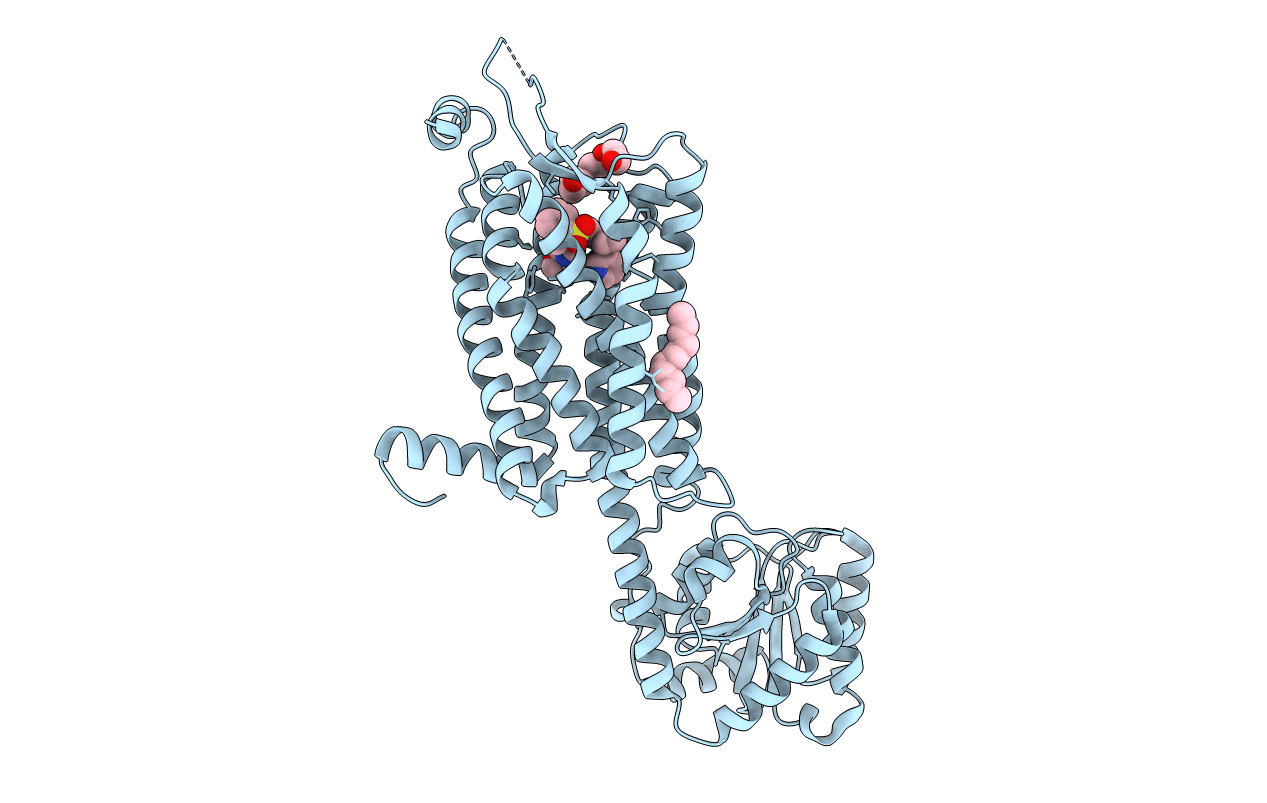
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2020-01-01 Classification: MEMBRANE PROTEIN Ligands: NT5, PG4, SO4, SOG, CL, PGW |
 |
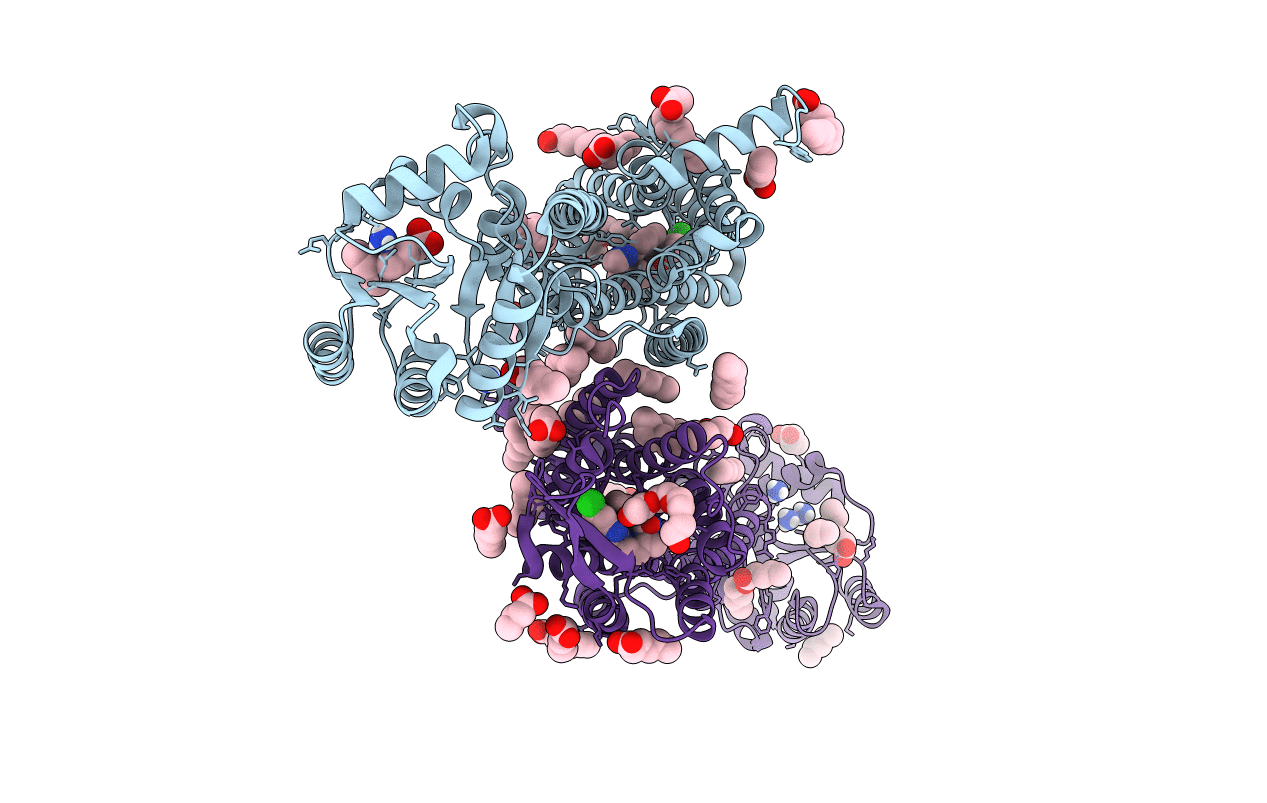
Crystal Structure Of The Orexin-2 Receptor In Complex With Empa At 2.74 A Resolution
Organism: Homo sapiens, Pyrococcus abyssi ge5
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2020-01-01 Classification: MEMBRANE PROTEIN Ligands: 7MA, PG4, OLA |
 |
Crystal Structure Of The Orexin-2 Receptor In Complex With Suvorexant At 2.76 A Resolution
Organism: Homo sapiens, Pyrococcus abyssi ge5
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2020-01-01 Classification: MEMBRANE PROTEIN Ligands: OLA, SUV, NH4, PG4 |

