Search Count: 26
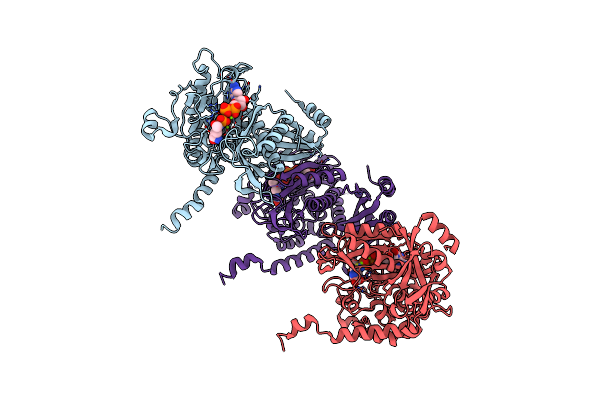 |
Structure Of Paenibacillus Polymyxa Gs Bound To Met-Sox-P-Adp (Transition State Complex) To 1.98 Angstom
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2022-06-29 Classification: LIGASE Ligands: ADP, P3S, MG |
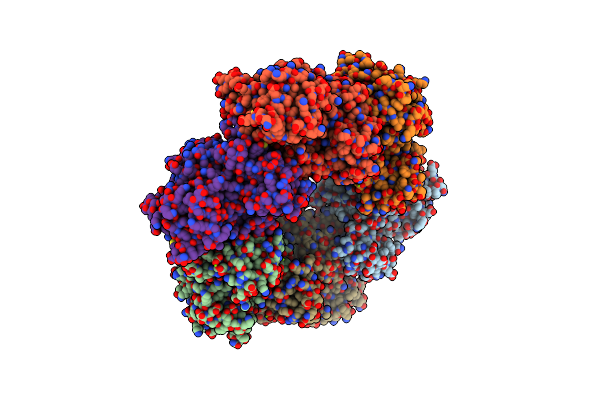 |
Crystal Structure Of S. Aureus Glutamine Synthetase In Met-Sox-P/Adp Transition State Complex
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2022-06-29 Classification: LIGASE Ligands: ADP, P3S, MG, SO4 |
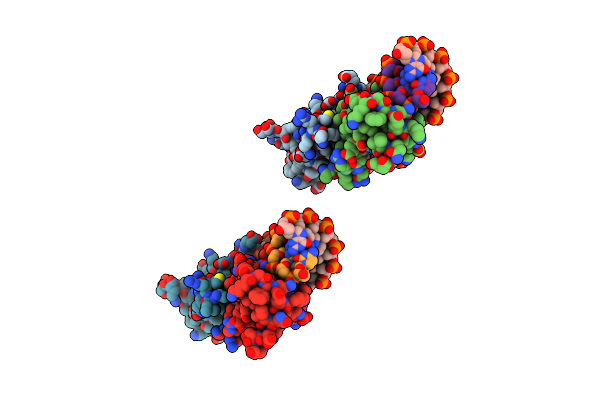 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-06-29 Classification: DNA BINDING PROTEIN/DNA Ligands: CA |
 |
Organism: Listeria monocytogenes, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2022-06-29 Classification: DNA BINDING PROTEIN/DNA |
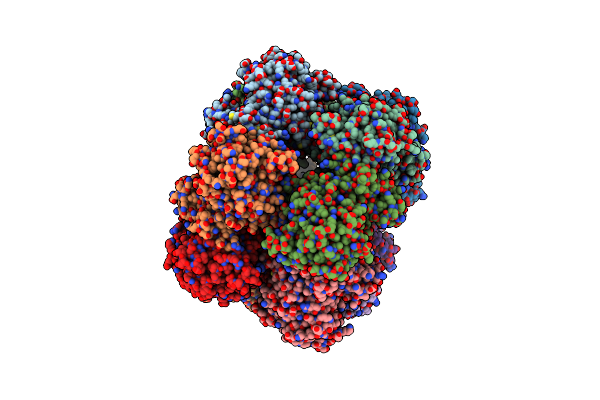 |
Crystal Structure Of The Listeria Monocytogenes Gs-Met-Sox-P- Adp Complex To 3.5 Angstrom
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2022-06-29 Classification: LIGASE/INHIBITOR Ligands: ADP, P3S |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE Ligands: MG, GLN |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE |
 |
Organism: Listeria monocytogenes
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE Ligands: MG, GLN |
 |
Organism: Paenibacillus polymyxa
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE Ligands: MG, GLN |
 |
Organism: Paenibacillus polymyxa
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE Ligands: MG, GLN |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE Ligands: MG, GLN |
 |
Organism: Paenibacillus polymyxa
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN Ligands: MG |
 |
Organism: Listeria monocytogenes
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN Ligands: MG |
 |
Crystal Structure Of Mura From Clostridium Difficile In The Presence Of Udp-N-Acetyl-Alpha-D-Muramic Acid With Modified Cys116 (S-[(1S)-1-Carboxy-1-(Phosphonooxy)Ethyl]-L-Cysteine)
Organism: Peptoclostridium difficile (strain 630)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-11-27 Classification: TRANSFERASE Ligands: EPZ, EDO |
 |
Crystal Structure Of Mura From Clostridium Difficile, Mutation C116D, N The Presence Of Udp-N-Acetylmuramic Acid
Organism: Peptoclostridium difficile (strain 630)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-11-27 Classification: TRANSFERASE Ligands: EDO, EPZ |
 |
Crystal Structure Of Mura From Clostridium Difficile, Mutant C116S, In The Presence Of Uridine-Diphosphate-N-Acetylglucosamine
Organism: Peptoclostridium difficile (strain 630)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-11-27 Classification: TRANSFERASE Ligands: EDO, UD1 |
 |
Crystal Structure Of Mura From Clostridium Difficile, Mutation C116S, In The Presence Of Uridine-Diphosphate-2(N-Acetylglucosaminyl) Butyric Acid
Organism: Peptoclostridium difficile (strain 630)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-11-27 Classification: TRANSFERASE Ligands: EPU, EDO, NA |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-04-24 Classification: LYASE Ligands: ZN, CL |
 |
X-Ray Structure Of A Glucosamine N-Acetyltransferase From Clostridium Acetobutylicum, Apo Form, Ph 5
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / lmg 5710 / vkm b-1787)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-07-06 Classification: TRANSFERASE Ligands: ACO, COA, EDO |
 |
X-Ray Structure Of A Glucosamine N-Acetyltransferase From Clostridium Acetobutylicum, Apo Form, Ph 8
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / lmg 5710 / vkm b-1787)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-07-06 Classification: TRANSFERASE Ligands: COA, ACO, EDO, PO4 |

