Search Count: 47
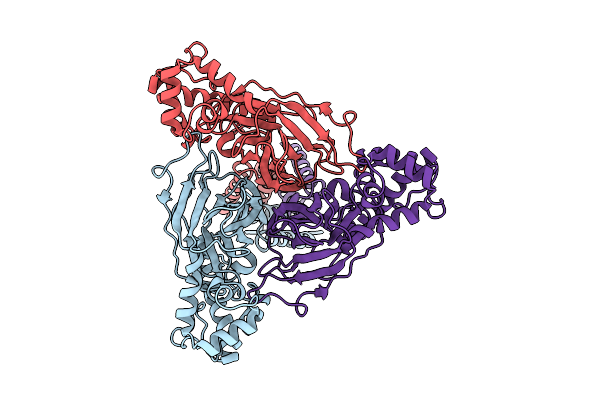 |
Organism: Scolopendra mutilans
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: MEMBRANE PROTEIN |
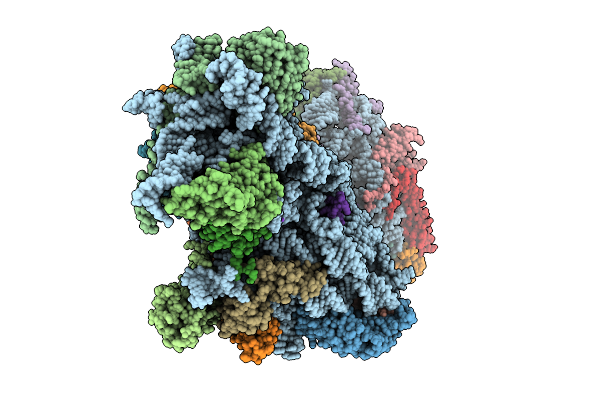 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: RIBOSOME Ligands: K, MG, ZN |
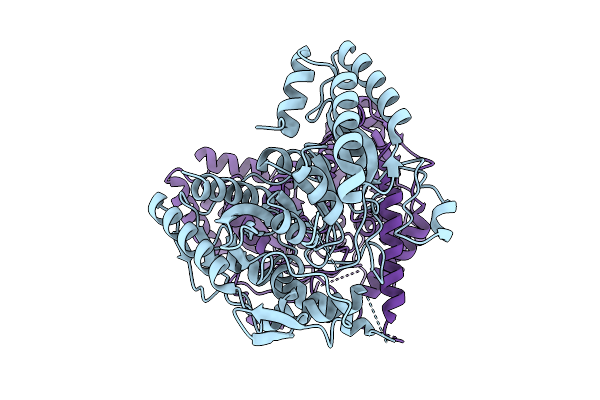 |
Organism: Sphingomonas sp. tpd3009
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE |
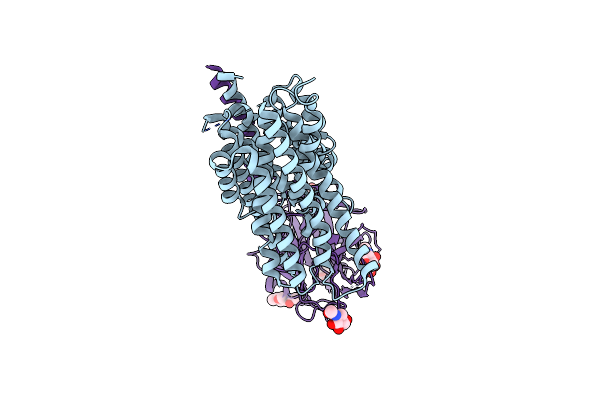 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: MEMBRANE PROTEIN Ligands: NAG |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: RIBOSOME Ligands: MG |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-04-19 Classification: CARBOHYDRATE Ligands: UNL |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: C8X |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: EDO |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: J1K |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: J1K |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: J1K |
 |
Cocrystallization Of Escherichia Coli Dihydrofolate Reductase (Dhfr) And Its Pyrrolo[3,2-F]Quinazoline Inhibitor.
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2022-04-27 Classification: ANTIBIOTIC Ligands: D4C, GOL |
 |
Crystal Structure Of A Novel Alpha/Beta Hydrolase In Apo Form In Complex With Citrate
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: CIT, MPD, SO4 |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: PGE |
 |
The Mutant Of Bifunctional Thioesterase Catalyzing Epimerization And Cyclization
Organism: Streptomyces abietis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-01-26 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-08-25 Classification: HYDROLASE |
 |
The Functional Characterization And Crystal Structure Of The Bifunctional Thioesterase Catalyzing Epimerization And Cyclization
Organism: Streptomyces abietis
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2021-08-18 Classification: BIOSYNTHETIC PROTEIN |

