Search Count: 31
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-04-17 Classification: PEPTIDE BINDING PROTEIN/ANTIBIOTIC |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2024-02-07 Classification: TRANSPORT PROTEIN/ANTIBIOTIC |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-02-07 Classification: PEPTIDE BINDING PROTEIN/ANTIBIOTIC |
 |
Crystal Structure Of The Substrate-Binding Protein Yeja In Complex With Peptide Fragment
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-09-20 Classification: TRANSPORT PROTEIN Ligands: GOL, CL, MG |
 |
The Crystal Structure Of Tfua Involved In Peptide Backbone Thioamidation From Methanosarcina Acetivorans
Organism: Methanosarcina acetivorans
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-03-17 Classification: BIOSYNTHETIC PROTEIN |
 |
The Structure Of A Coa-Dependent Acyl-Homoserine Lactone Synthase, Rpai, With The Adduct Of Sah And P-Coumaroyl Coa
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-05-13 Classification: TRANSFERASE Ligands: COA, U4Y |
 |
Organism: Mesorhizobium sp. ors 3359
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-05-13 Classification: TRANSFERASE |
 |
Structure Of Ycao Enzyme From Methanocaldococcus Jannaschii In Complex With Atp
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-01-08 Classification: BIOSYNTHETIC PROTEIN Ligands: ATP |
 |
Structure Of Ycao Enzyme From Methanocaldococcus Jannaschii In Complex With Peptide
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440), Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-11-06 Classification: BIOSYNTHETIC PROTEIN Ligands: ATP, MG |
 |
The Structure Of Microcin C7 Biosynthetic Enzyme Mccb In Complex With N-Formylated Mcca
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-05-22 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, MG, POP, ND7 |
 |
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 10
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FXD |
 |
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 11
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FXS |
 |
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 12
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FYD, HIS |
 |
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 13
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FY7 |
 |
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 14
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FY4 |
 |
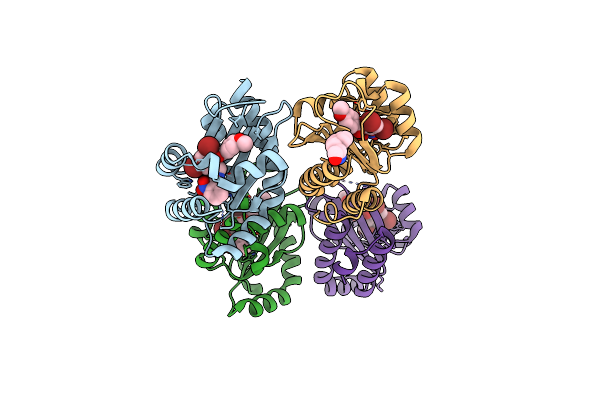
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 15
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: TX7 |
 |
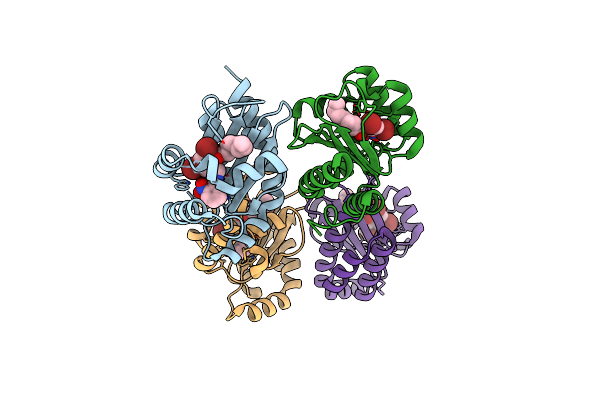
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 16
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FXJ, PHE |
 |
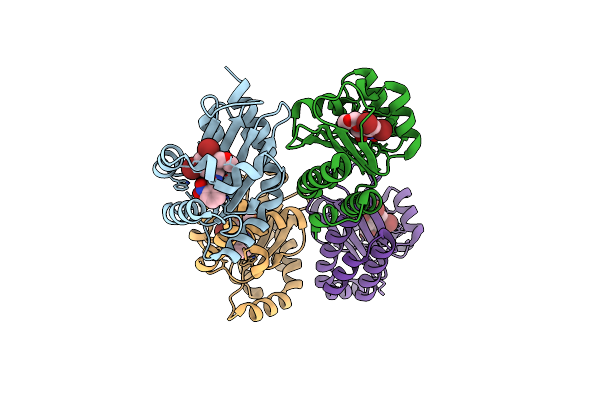
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 17
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FX7 |
 |
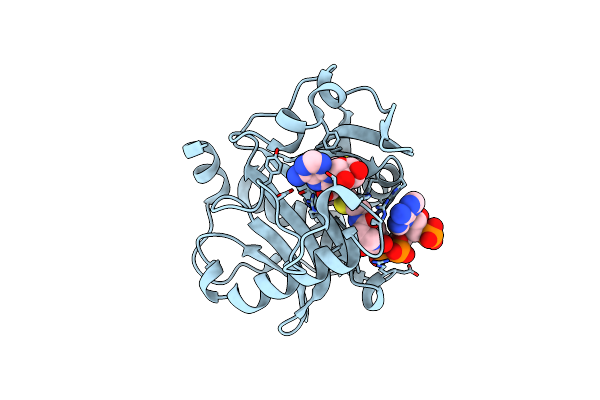
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 19
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FY1 |
 |
The Structure Of A Coa-Dependent Acyl-Homoserine Lactone Synthase, Bjai, With Sam And Isopentyl-Coa
Organism: Bradyrhizobium japonicum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: SAM, A1S |

