Search Count: 222
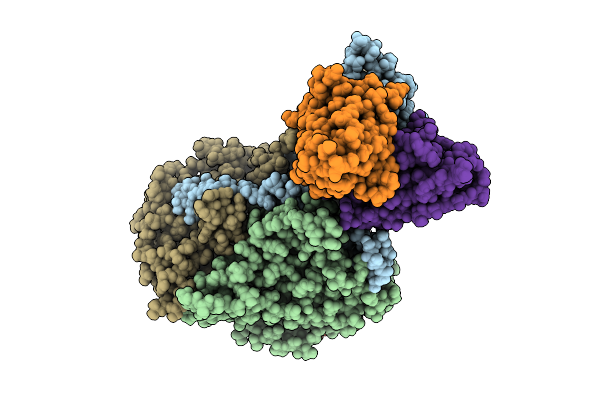 |
Organism: Poliovirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRUS Ligands: PLM |
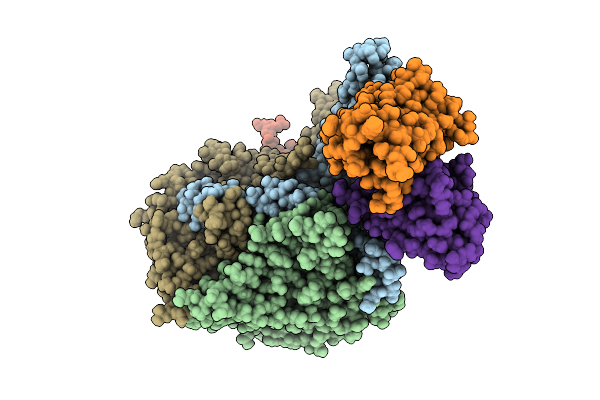 |
Organism: Poliovirus 3, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRUS Ligands: PLM |
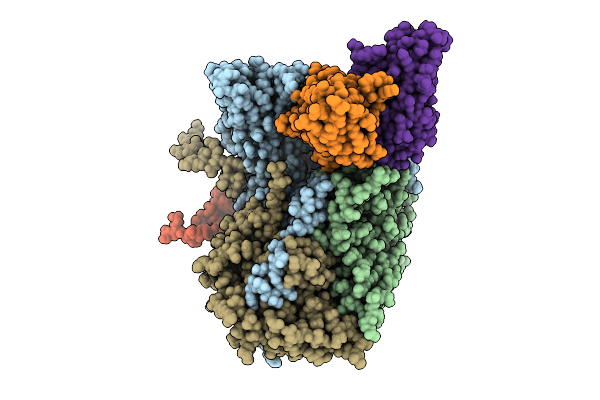 |
Organism: Poliovirus 3, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRUS Ligands: PLM |
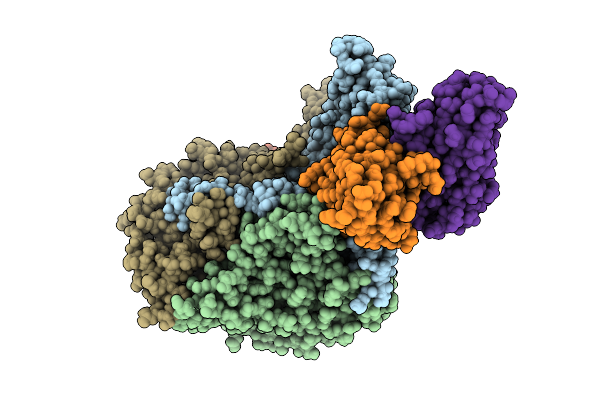 |
Organism: Human poliovirus 1 strain sabin, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRUS Ligands: PLM |
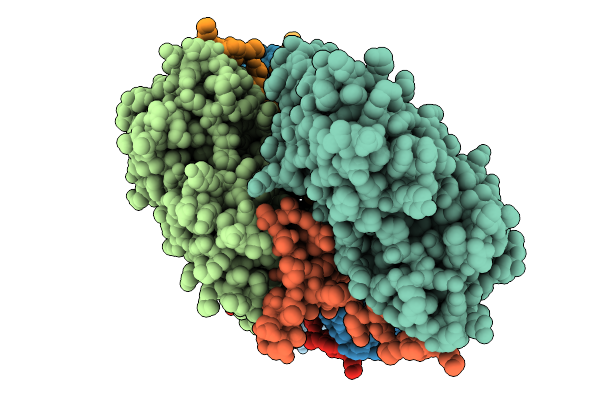 |
Atomic Structure Of Vibrio Effector Fragment Vopv Bound To Beta-Cytoplasmic/Gamma1-Cytoplasmic F-Actin
Organism: Homo sapiens, Vibrio parahaemolyticus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: STRUCTURAL PROTEIN Ligands: MG, ANP |
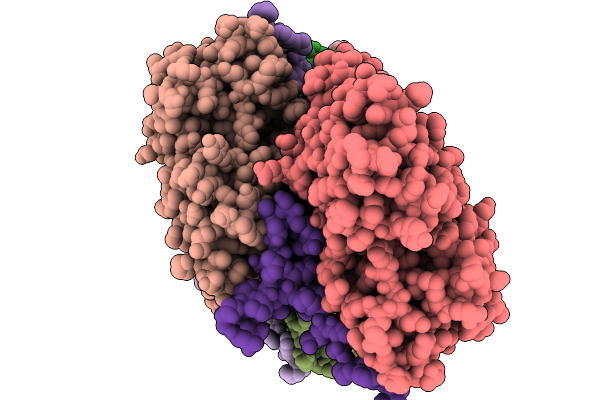 |
Cryo-Em Structure Of Vibrio Effector Vopv Fragment Bound To Skeletal Alpha F-Actin
Organism: Oryctolagus cuniculus, Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: STRUCTURAL PROTEIN Ligands: MG, ANP |
 |
Organism: Caldicellulosiruptor sp.
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Crystal Structure Of Beta-Glucosidase Cabgl Mutant E163Q In Complex With Glucose
Organism: Caldicellulosiruptor sp.
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: HYDROLASE Ligands: GOL, BGC, SO4 |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/DNA |
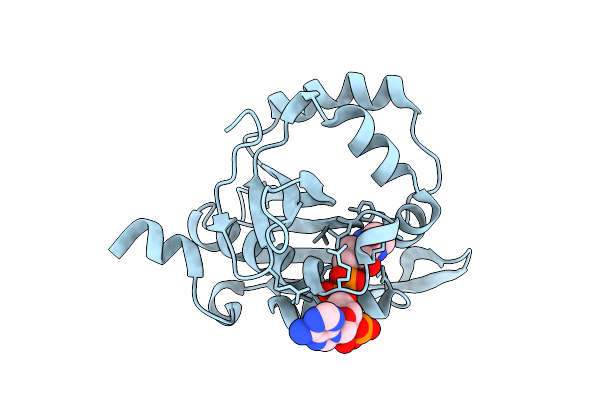 |
Iterative Acetyltransferase On Lasso Peptides From Actinomycetes In Complex With Coa
Organism: Actinosynnema mirum (strain atcc 29888 / dsm 43827 / jcm 3225 / nbrc 14064 / ncimb 13271 / nrrl b-12336 / imru 3971 / 101)
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: COA |
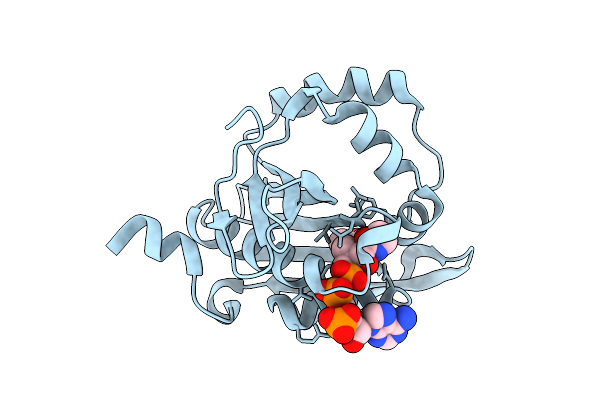 |
Iterative Acetyltransferase On Lasso Peptides From Actinomycetes In Complex With Accoa
Organism: Actinosynnema mirum (strain atcc 29888 / dsm 43827 / jcm 3225 / nbrc 14064 / ncimb 13271 / nrrl b-12336 / imru 3971 / 101)
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: ACO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: PROTEIN FIBRIL Ligands: BDP |
 |
Drimenyl Diphosphate Synthase Ssdms_F248A&D303E From Streptomyces Showdoensis (Apo)
Organism: Streptomyces showdoensis
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2025-05-21 Classification: BIOSYNTHETIC PROTEIN |
 |
Drimenyl Diphosphate Synthase Ssdms_F248Y&D303E From Streptomyces Showdoensis (Apo)
Organism: Streptomyces showdoensis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-05-21 Classification: BIOSYNTHETIC PROTEIN |
 |
Drimenyl Diphosphate Synthase Ssdms_Y505I&D303A From Streptomyces Showdoensis (Apo)
Organism: Streptomyces showdoensis
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2025-05-21 Classification: BIOSYNTHETIC PROTEIN |
 |
Drimenyl Diphosphate Synthase Ssdms_F248A&D303E From Streptomyces Showdoensis In Complex With Farnesyl Diphosphate (Fpp) And Mg2+
Organism: Streptomyces showdoensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-21 Classification: BIOSYNTHETIC PROTEIN Ligands: FPP, MG |

