Search Count: 352
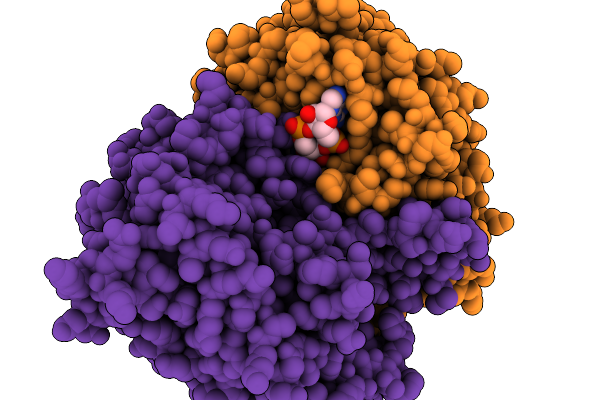 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE Ligands: 4BW |
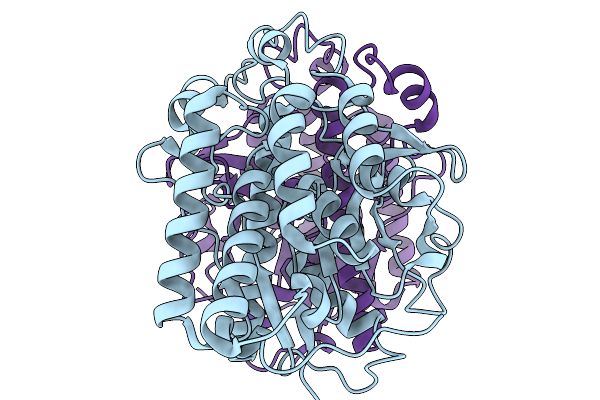 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE |
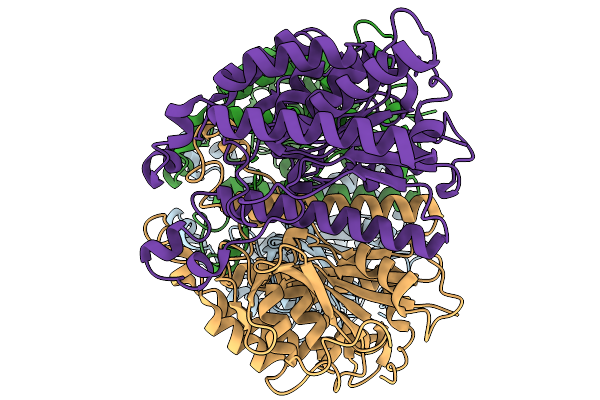 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE |
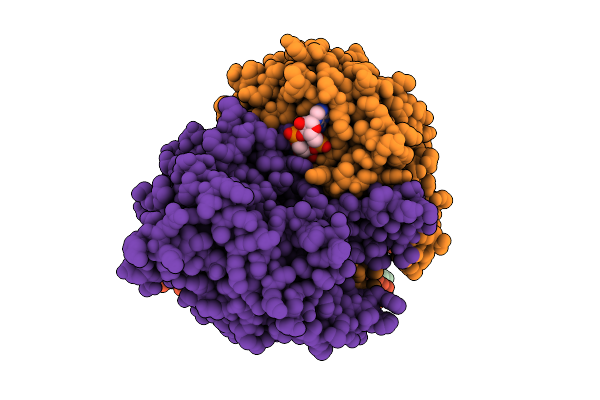 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: HYDROLASE Ligands: 4BW |
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: CO, 92X |
 |
Organism: Photorhabdus asymbiotica
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: AKG, SO4, GOL, PEG, FE |
 |
Pasi From Photorhabdus Asymbiotica Bound To Vanadyl, Succinate, And 5-Amino-6-Hydroxy-Octanosyl Acid 2-Phosphate
Organism: Photorhabdus asymbiotica
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: A1BIK, GOL, SIN, CL, V |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Zea Mays 3-Phosphoglycerate Dehydrogenase S282L Mutant
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE |
 |
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: CO, A1L7D |
 |
The Crystal Structure Of The Ha1 Domain Of Hemagglutinin From A/Shanghai/02/2013 (H7N9) Bound To H7-235 Fab
Organism: Influenza a virus (a/shanghai/02/2013(h7n9)), Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: IMMUNE SYSTEM Ligands: NAG, SIA |
 |
Organism: Arachis hypogaea
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: PLANT PROTEIN Ligands: ZN, CDL, 3PE, HEM, HEC, FES, PC1 |
 |
Mycobacterium Tuberculosis Pks13 Acyltransferase Serine Converted To Beta-Lactam Form By Cec215 Via Sufex Reaction
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: ANTIBIOTIC Ligands: SO4, CL, PG4, EDO, DMS, GOL, PEG |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: ANTIBIOTIC Ligands: DMS, SO4, PE5, EDO, PEG, GOL, CL, NA, P33 |
 |
Crystal Structure Of Ppargamma Ligand Binding Domain (Lbd) In Complex With Ncor1 Corepressor Peptide And Inverse Agonist Fx-909
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: NUCLEAR PROTEIN Ligands: A1CAA |
 |
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cec215
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 1PE, SO4, A1ATV |
 |
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cmx410
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE/TRANSFERSE INHIBITOR Ligands: 1PE, SO4, A1ATW |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE Ligands: 1PE, SO4 |
 |
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cmx410 - Reaction Product
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE Ligands: SO4, 1PE, A1AVL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: CA |

