Search Count: 43
 |
Crystal Structure Of The Catalase-Peroxidase From B. Pseudomallei With Maltose Bound
Organism: Burkholderia pseudomallei 1710b
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-07-04 Classification: OXIDOREDUCTASE Ligands: HEM, NA, CL, OXY, PO4, MPD |
 |
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2016-09-07 Classification: OXIDOREDUCTASE Ligands: HEM, NA, CL, OXY, MPD, MRD, ADP |
 |
Crystal Structure Of Catalase-Peroxidase Katg With Isonicotinic Acid Hydrazide And Amp Bound
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2016-09-07 Classification: OXIDOREDUCTASE Ligands: HEM, NA, CL, OXY, PO4, MRD, NIZ, MPD, AMP |
 |
Crystal Structure Of The S324T Variant Of Burkholderia Pseudomallei Katg With Isonicotinic Acid Hydrazide Bound
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-09-07 Classification: OXIDOREDUCTASE Ligands: HEM, NA, CL, OXY, PO4, MPD, NIZ |
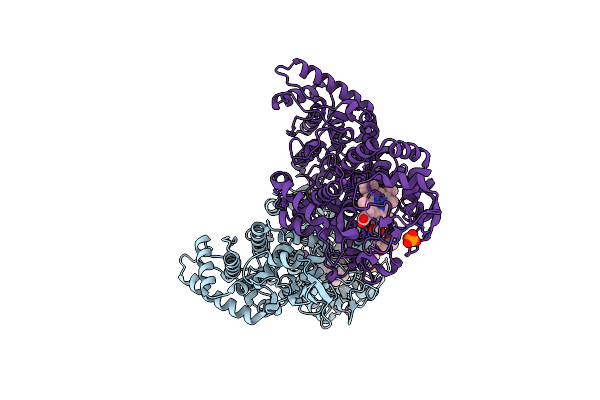 |
Crystal Structure Of The E198A Variant Of Catalase-Peroxidase Katg Of Burkholderia Pseudomallei
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-09-07 Classification: OXIDOREDUCTASE Ligands: HEM, NA, OXY, PO4, MRD, MPD |
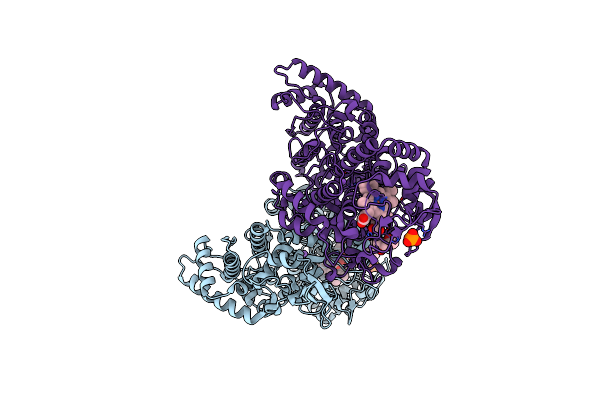 |
Crystal Structure Of The E198A Variant Of Burkholderia Pseudomallei Catalase-Peroxidase Katg With Inh
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-09-07 Classification: OXIDOREDUCTASE Ligands: HEM, NA, CL, OXY, PO4, MPD |
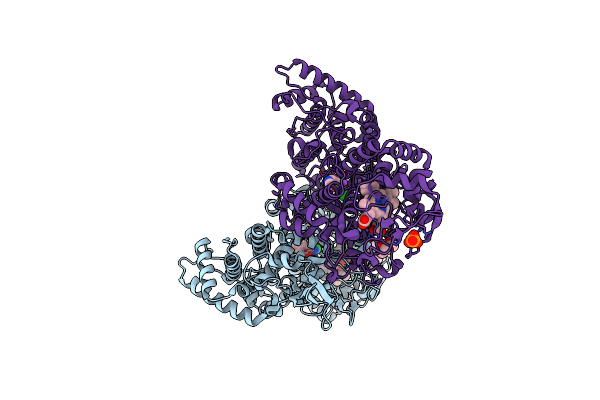 |
Crystal Structure Of B. Pseudomallei Katg With Isonicotinic Acid Hydrazide Bound
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-08-31 Classification: OXIDOREDUCTASE Ligands: HEM, NA, CL, OXY, MPD, PO4, NIZ |
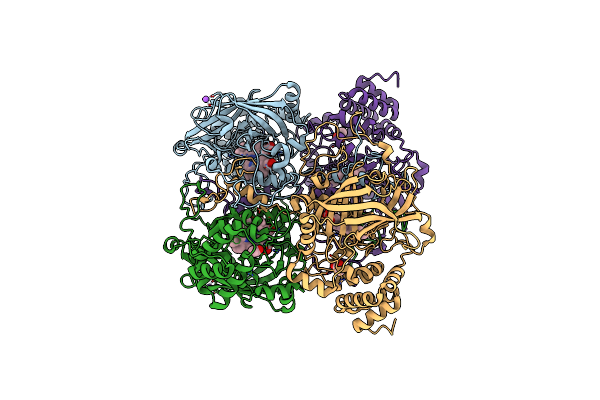 |
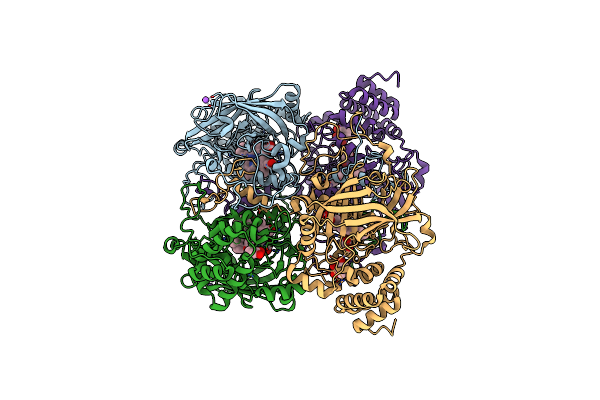
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE Ligands: HEM, CL, NA, ACT |
 |
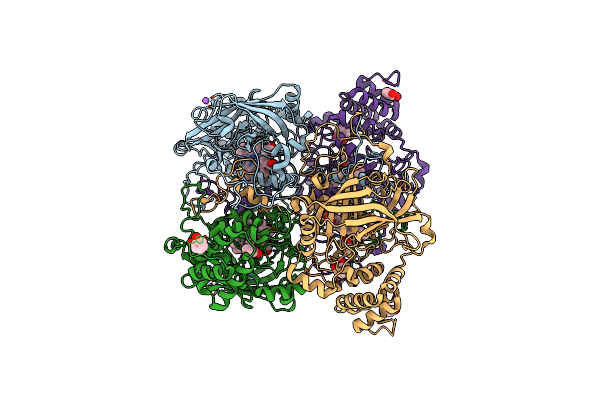
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE Ligands: HEM, PYG, CL, NA |
 |
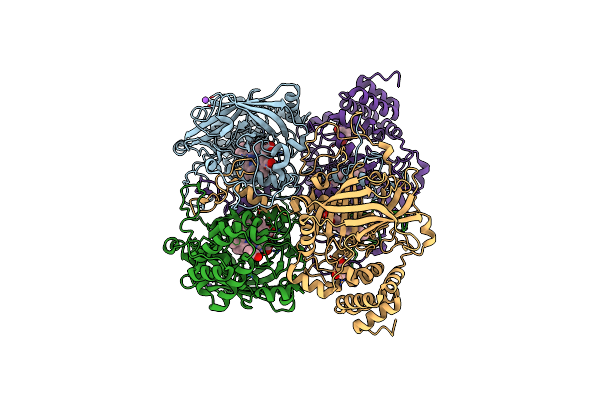
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE Ligands: HEM, CAQ, CL, NA |
 |
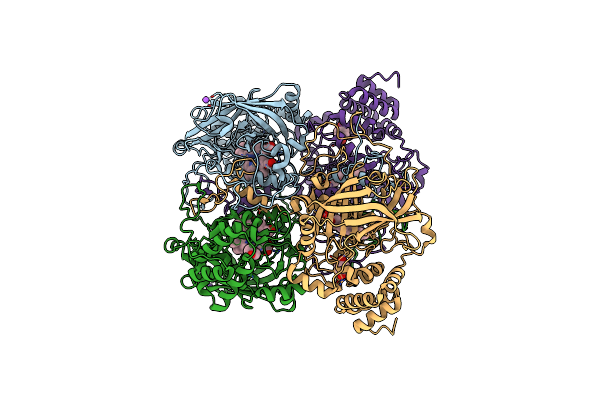
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE Ligands: HEM, RCO, CL, NA |
 |
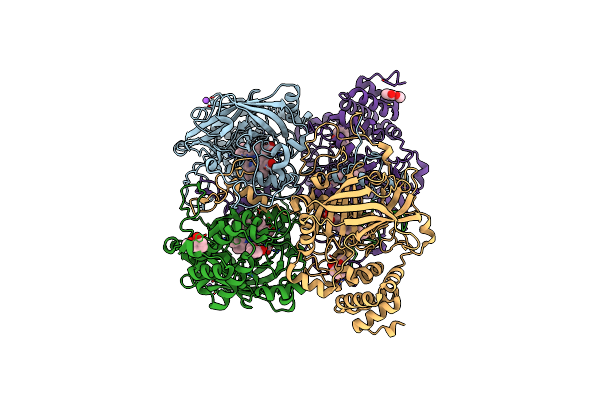
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE Ligands: HEM, HQE, CL, NA |
 |
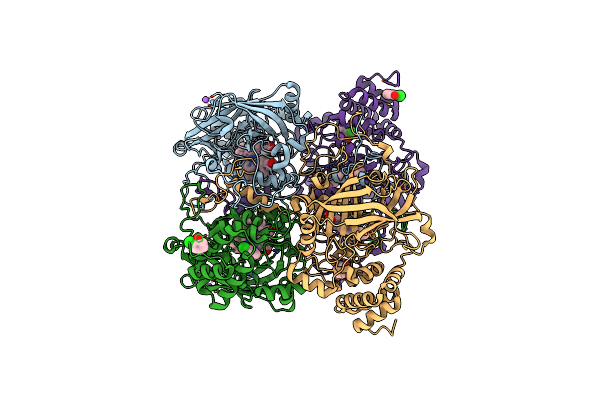
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE Ligands: HEM, JZ3, CL, NA |
 |
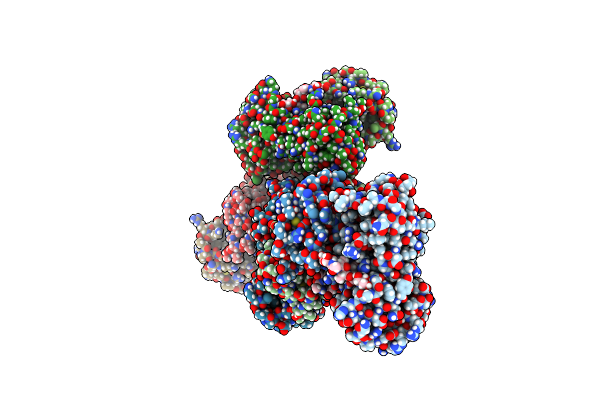
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE Ligands: HEM, 2CH, CL, NA |
 |
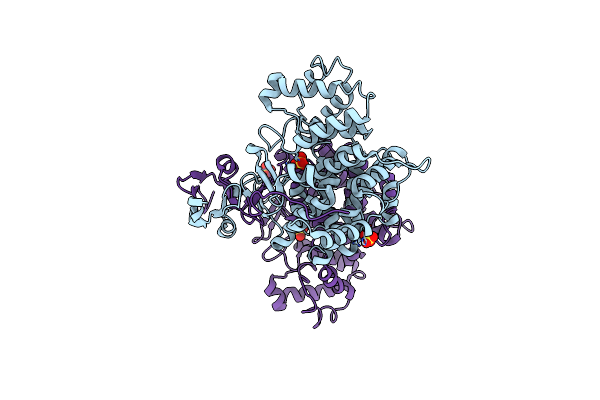
Ampr Effector Binding Domain From Citrobacter Freundii Bound To Udp-Murnac-Pentapeptide
Organism: Citrobacter freundii atcc 8090 = mtcc 1658
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-12-17 Classification: TRANSCRIPTION Ligands: GOL, MUB |
 |
Structural Determination Of The A50T:S279G:S280K:V281K:K282E:H283N Variant Of Citrate Synthase From E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-07-17 Classification: TRANSFERASE Ligands: SO4 |
 |
Structural Determination Of The A50T:S279G:S280K:V281K:K282E:H283N Variant Of Citrate Synthase From E. Coli Complexed With S-Carboxymethyl-Coa
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-07-17 Classification: TRANSFERASE Ligands: SO4, CMC |
 |
Structural Determination Of The A50T:S279G:S280K:V281K:K282E:H283N Variant Of Citrate Synthase From E. Coli Complexed With Nadh
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-07-17 Classification: TRANSFERASE Ligands: SO4, NAI |
 |
Structural Determination Of The A50T:S279G:S280K:V281K:K282E:H283N Variant Of Citrate Synthase From E. Coli Complexed With Oxaloacetate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-07-17 Classification: TRANSFERASE Ligands: OAA, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2011-10-12 Classification: OXIDOREDUCTASE Ligands: HEM |

