Search Count: 44
 |
Organism: Bifidobacterium longum
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Bifidobacterium longum
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Bifidobacterium longum
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: HYDROLASE Ligands: ZN |
 |
Cryo-Em Structure Of Bl_Man38A Nucleophile Mutant In Complex With Mannose At 2.7 A
Organism: Bifidobacterium longum
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: HYDROLASE Ligands: ZN, MAN |
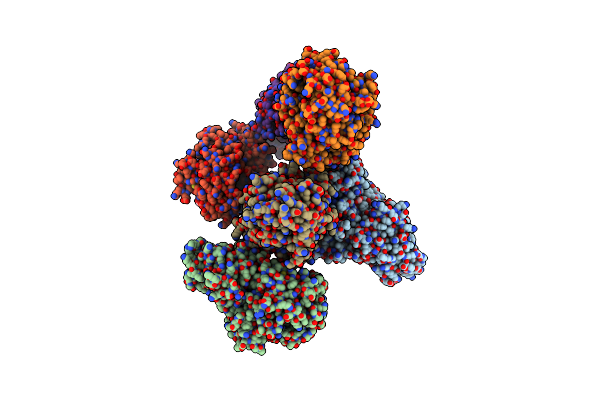 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2021-09-08 Classification: HYDROLASE Ligands: GOL |
 |
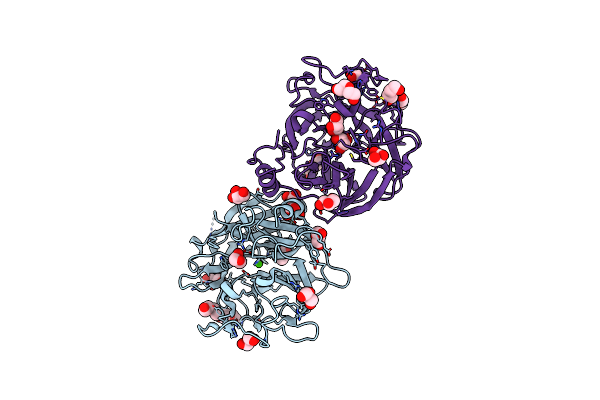
Crystal Structure Of A Beta-Helix Domain Retrieved From Capybara Gut Metagenome
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-09-08 Classification: UNKNOWN FUNCTION Ligands: CA |
 |
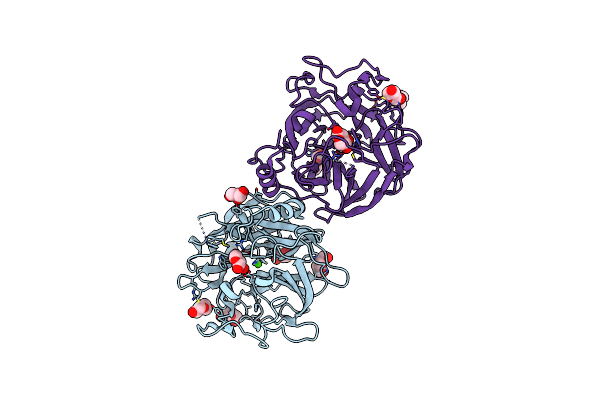
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: CA, GOL, PEG |
 |
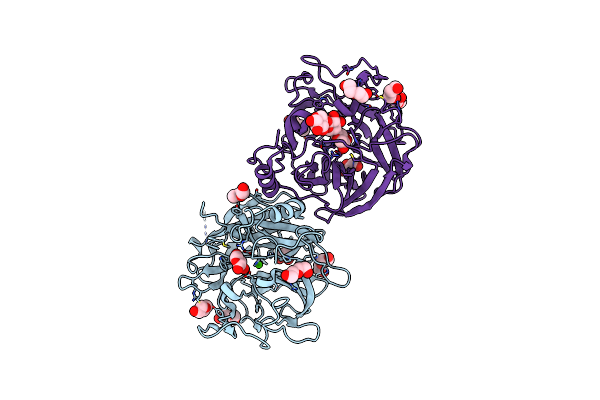
Crystal Structure Of The Gh43_1 Enzyme From Xanthomonas Citri Complexed With Xylose
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: CA, XYP, PEG, GOL |
 |
Crystal Structure Of The Gh43_1 Enzyme From Xanthomonas Citri Complexed With Xylotriose
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: CA, XYP, PEG, GOL |
 |
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2020-07-22 Classification: HYDROLASE |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase From Amycolatopsis Mediterranei (Amgh128_I)
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: GOL, NA |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaritriose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-05-20 Classification: HYDROLASE |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase (E199A Mutant) From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaripentaose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: BGC, ZN, PEG |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase (E102A Mutant) From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaripentaose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase (E102A Mutant) From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaritriose And Laminaribiose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: ZN, PEG |
 |
Crystal Structure Of A Gh128 (Subgroup Ii) Endo-Beta-1,3-Glucanase From Pseudomonas Viridiflava (Pvgh128_Ii)
Organism: Pseudomonas viridiflava
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Crystal Structure Of A Gh128 (Subgroup Ii) Endo-Beta-1,3-Glucanase From Pseudomonas Viridiflava (Pvgh128_Ii) In Complex With Laminaritriose
Organism: Pseudomonas viridiflava
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of A Gh128 (Subgroup Ii) Endo-Beta-1,3-Glucanase From Sorangium Cellulosum (Scgh128_Ii)
Organism: Sorangium cellulosum so ce56
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: TRS, EPE, CA, CL |
 |
Crystal Structure Of A Gh128 (Subgroup Iii) Curdlan-Specific Exo-Beta-1,3-Glucanase From Blastomyces Gilchristii (Bggh128_Iii)
Organism: Blastomyces gilchristii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-05-20 Classification: HYDROLASE |
 |
Crystal Structure Of A Gh128 (Subgroup Iii) Curdlan-Specific Exo-Beta-1,3-Glucanase From Blastomyces Gilchristii (Bggh128_Iii) In Complex With Glucose
Organism: Blastomyces gilchristii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: BGC |

