Search Count: 12
 |
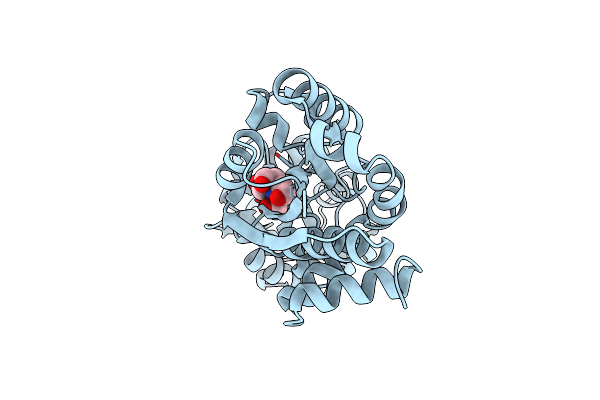
The Putative Substrate Binding Domain Of Abc-Type Transporter From Agrobacterium Tumefaciens In Open Conformation
Organism: Agrobacterium fabrum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-12-11 Classification: TRANSPORT PROTEIN |
 |
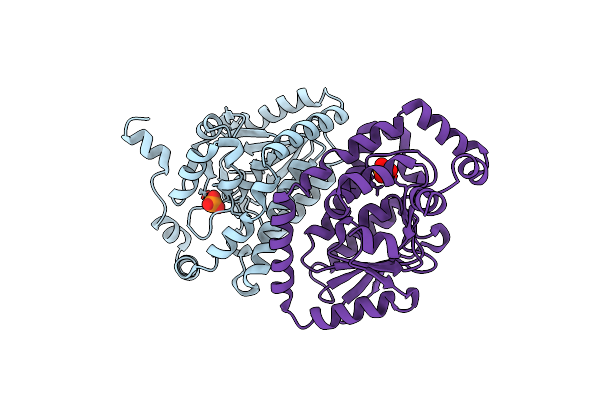
Crystal Structure Of Abc Transporter Substrate Binding Protein Prox From Agrobacterium Tumefaciens Cocrystalized With Btb
Organism: Agrobacterium fabrum
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2013-11-27 Classification: TRANSPORT PROTEIN Ligands: BTB, CL |
 |
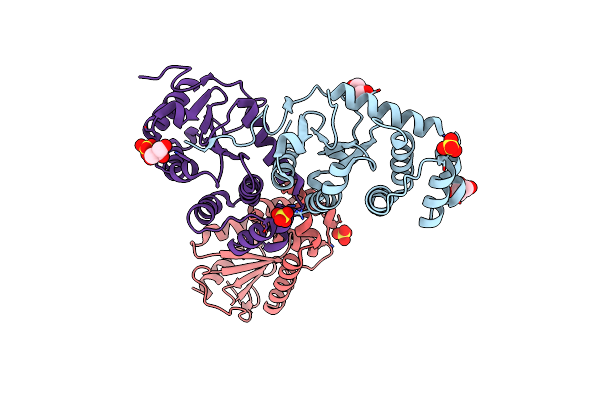
Crystal Structure Of Putative Enoyl-Coa Hydratase/Isomerase From Novosphingobium Aromaticivorans Dsm 12444
Organism: Novosphingobium aromaticivorans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-05-29 Classification: ISOMERASE Ligands: UNL |
 |
Crystal Structure Of Disulfide Bond Oxidoreductase Dsba1 From Legionella Pneumophila
Organism: Legionella pneumophila subsp. pneumophila
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2013-04-24 Classification: OXIDOREDUCTASE Ligands: SO4, GOL |
 |
Crystal Structure Of C103A Mutant Of Dj-1 Superfamily Protein Stm1931 From Salmonella Typhimurium
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-04-24 Classification: UNKNOWN FUNCTION Ligands: ZN |
 |
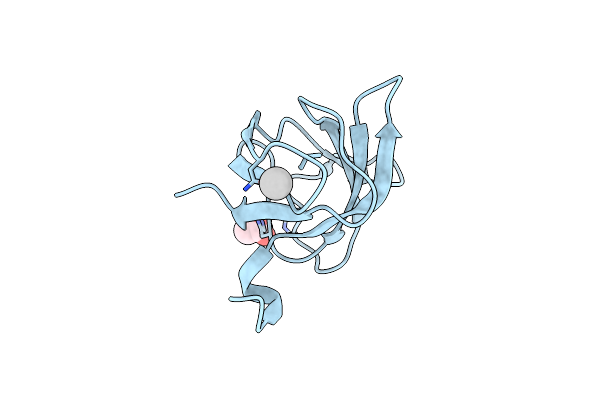
Crystal Structure Of The Selenomethionine Variant Of The C-Terminal Domain Of Geobacillus Thermoleovorans Putative U32 Peptidase
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2012-11-14 Classification: UNKNOWN FUNCTION Ligands: SO4 |
 |
Crystal Structure Of The C-Terminal Domain Of Geobacillus Thermoleovorans Putative U32 Peptidase
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2012-11-14 Classification: UNKNOWN FUNCTION Ligands: UNX, ACT |
 |
Crystal Structure Of The Luciferase-Like Monooxygenase From Bacillus Cereus Atcc 10987.
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-05-11 Classification: Structural Genomics, Unknown function Ligands: SO4 |
 |
Crystal Structure Of Putative 2,3-Dihydroxybenzoate-Specific Isochorismate Synthase, Dhbc From Bacillus Anthracis.
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-10-20 Classification: ISOMERASE Ligands: SO4, 15P, GOL |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2010-06-16 Classification: structural genomics, unknown function Ligands: CL |
 |
1.95 Angstrom Crystal Structure Of Coce/Nond Family Hydrolase (Sacol2612) From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-08-18 Classification: HYDROLASE Ligands: NI, CL, PLM |
 |
Crystal Structure Of Sacol2612 - Coce/Nond Family Hydrolase From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2009-08-11 Classification: HYDROLASE Ligands: CL, PLM, NI |

