Search Count: 13
 |
Crystal Structure Of The Microbial Rhodopsin From Sphingomonas Paucimobilis (Spar)
Organism: Sphingomonas paucimobilis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-05-03 Classification: MEMBRANE PROTEIN Ligands: LFA |
 |
Crystal Structure Of Cold-Active Esterase Pmgl3 From Permafrost Metagenomic Library
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-11-03 Classification: HYDROLASE |
 |
Crystal Structure Of C173S Mutation In The Pmgl2 Esterase From Permafrost Metagenomic Library
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: MG, PG6 |
 |
Crystal Structure Of The Pmgl2 Esterase From Permafrost Metagenomic Library
Organism: Permafrost metagenome
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: CL, MG |
 |
Crystal Structure Of The Pmgl2 Esterase (Point Mutant 1) From Permafrost Metagenomic Library
Organism: Permafrost metagenome
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: CL, PG6, MG |
 |
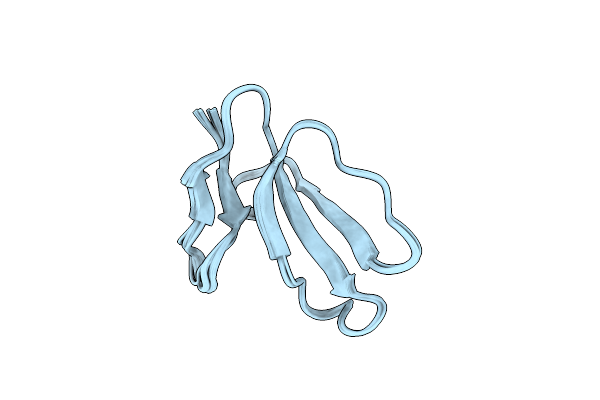
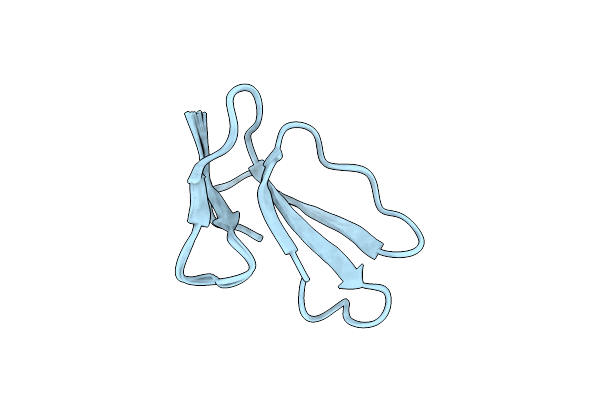
Organism: Naja oxiana
Method: SOLUTION NMR Release Date: 2018-04-04 Classification: TOXIN Ligands: HOH |
 |
 |
 |
 |
Crystal Structure Of Cold-Active Estarase From Psychrobacter Cryohalolentis K5T
Organism: Psychrobacter cryohalolentis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2015-01-07 Classification: HYDROLASE |
 |
Organism: Exiguobacterium sibiricum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-07-17 Classification: PROTON TRANSPORT Ligands: RET, LFA |
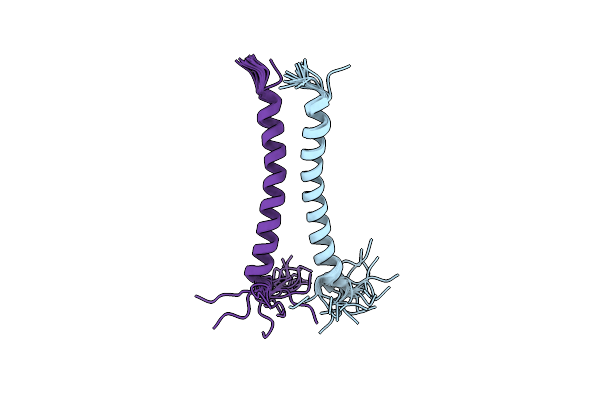 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2011-05-18 Classification: MEMBRANE PROTEIN |
 |

