Search Count: 19
 |
The Crystal Structure Of Cyp125Mrca, An Ancestrally Reconstructed Cyp125 Enzyme
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: LIPID BINDING PROTEIN Ligands: HEM, VD3, EDO, IPA, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: LIPID BINDING PROTEIN Ligands: A1LXL, HEM |
 |
Crystal Structure Of Phospholipase D (Pld) From Arcanobacterium Haemolyticum
Organism: Arcanobacterium haemolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: HYDROLASE Ligands: GOL, MG |
 |
Substrate Free Structure Of Cytochrome P450 Cyp105Q4 From Mycobacterium Marinum
Organism: Mycobacterium marinum atcc baa-535
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2024-03-27 Classification: OXIDOREDUCTASE Ligands: HEM, PGE |
 |
The Crystal Structure Of T252E Cyp199A4 Bound To 4-(Pyridin-2-Yl)Benzoic Acid
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2023-10-25 Classification: OXIDOREDUCTASE Ligands: CL, HEM, PQS |
 |
The Crystal Structure Of T252E Cyp199A4 Bound To 4-(Thiophen-3-Yl)Benzoic Acid
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2023-10-25 Classification: OXIDOREDUCTASE Ligands: EJD, HEM, CL |
 |
The Crystal Structure Of The T252E Mutant Of Cyp199A4 Bound To 4-Benzylbenzoic Acid
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2023-10-25 Classification: OXIDOREDUCTASE Ligands: IRJ, HEM, CL |
 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-08-30 Classification: OXIDOREDUCTASE Ligands: MH0, HX1, CL |
 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: HEM, JO4, CL |
 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: HEM, Z7Z, CL, MG |
 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: HEM, EH1, CL |
 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: HEM, 8ZU, CL |
 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: QDB, HEM, CL, MG |
 |
Crystal Structure Of Cyp142A3 From Mycobacterium Ulcerans Bound To Cholest-4-En-3-One
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-10-05 Classification: LIPID BINDING PROTEIN,OXIDOREDUCTASE Ligands: HEM, K2B, NA, ACT |
 |
X-Ray Crystal Structure Of Cyp142A3 From Mycobacterium Marinum In Complex With 4-Cholesten-3-One
Organism: Mycobacterium marinum
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-09-14 Classification: OXIDOREDUCTASE Ligands: K2B, HEM, ACT, NA |
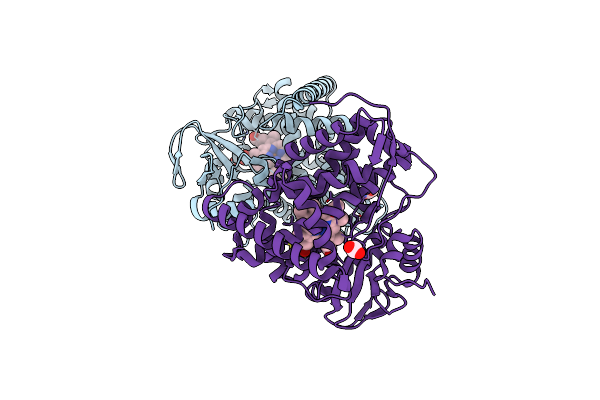 |
X-Ray Crystal Structure Of Substrate Free Cytochrome P450 Cyp142A3 From Mycobacterium Marinum
Organism: Mycobacterium marinum atcc baa-535
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-09-14 Classification: OXIDOREDUCTASE Ligands: HEM, ACT |
 |
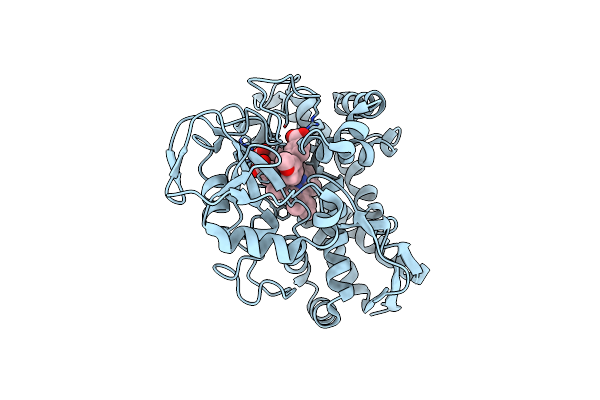
Organism: Rhodopseudomonas palustris (strain haa2)
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2022-06-29 Classification: OXIDOREDUCTASE Ligands: HEM, 8ZU, CL |
 |
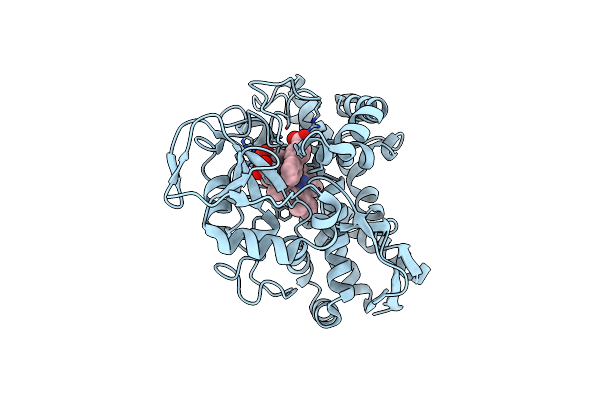
Organism: Rhodopseudomonas palustris (strain haa2)
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2022-03-09 Classification: OXIDOREDUCTASE Ligands: VLG, HEM, CL |
 |
Organism: Rhodopseudomonas palustris (strain haa2)
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2021-09-01 Classification: OXIDOREDUCTASE Ligands: Z7Z, HEM, CL |

