Search Count: 131
 |
Organism: Leishmania donovani
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: LIGASE |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-03-05 Classification: HYDROLASE Ligands: CL, NA, GOL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-03-05 Classification: HYDROLASE Ligands: GOL, CL |
 |
The Crystal Structure Of Full Length Tetramer Cysb From Klebsiella Aerogenes In Complex With N-Acetylserine
Organism: Klebsiella aerogenes
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-07-24 Classification: TRANSCRIPTION Ligands: SAC |
 |
The Crystal Structure Of Full Length Tetramer Cysb From Klebsiella Aerogenes In Complex With N-Acetylserine
Organism: Klebsiella
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-07-03 Classification: TRANSCRIPTION Ligands: SAC |
 |
The Structure Of Membrane-Active Antibiotic Cyclodecapeptide Gramicidin S In Complex With Urea
Organism: Brevibacillus brevis
Method: X-RAY DIFFRACTION Resolution:0.98 Å Release Date: 2024-03-06 Classification: ANTIBIOTIC Ligands: URE |
 |
X-Ray Structure Of The Ceue Homologue From Geobacillus Stearothermophilus - Apo Form.
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2023-07-12 Classification: METAL TRANSPORT Ligands: JEF, SO4 |
 |
X-Ray Structure Of The Ceue Homologue From Geobacillus Stearothermophilus - 5-Licam Siderophore Analogue Complex.
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2023-07-12 Classification: METAL TRANSPORT Ligands: FE, 5LC |
 |
X-Ray Structure Of The Ceue Homologue From Geobacillus Stearothermophilus - Azotochelin Complex.
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2023-07-12 Classification: METAL TRANSPORT Ligands: FE, 95B |
 |
X-Ray Structure Of The Ceue Homologue From Parageobacillus Thermoglucosidasius - Azotochelin Complex
Organism: Parageobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2023-07-12 Classification: METAL TRANSPORT Ligands: FE, 95B, NI, SO4 |
 |
X-Ray Structure Of The Ceue Homologue From Parageobacillus Thermoglucosidasius - 5Licam Complex.
Organism: Parageobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-07-12 Classification: METAL TRANSPORT Ligands: FE, 5LC, NI, SO4 |
 |
X-Ray Structure Of The Ceue Homologue From Parageobacillus Thermoglucosidasius - Apo Form
Organism: Parageobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2023-07-12 Classification: METAL TRANSPORT Ligands: NI, SO4 |
 |
Crystal Structure Of The Peptide Binding Protein, Oppa, From Bacillus Subtilis In Complex With A Phre-Derived Pentapeptide
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
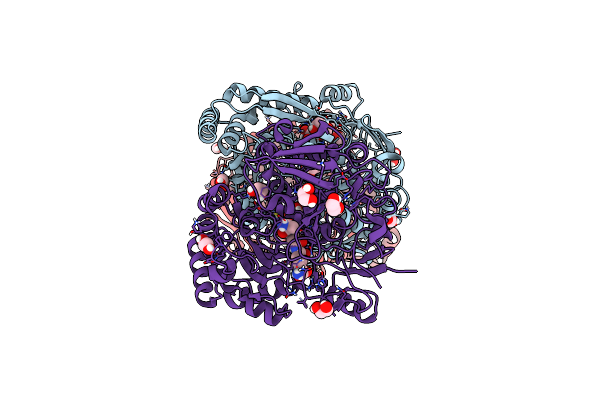
Crystal Structure Of The Peptide Binding Protein, Oppa, From Bacillus Subtilis In Complex With An Endogenous Tetrapeptide
Organism: Bacillus subtilis subsp. subtilis str. 168, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN |
 |
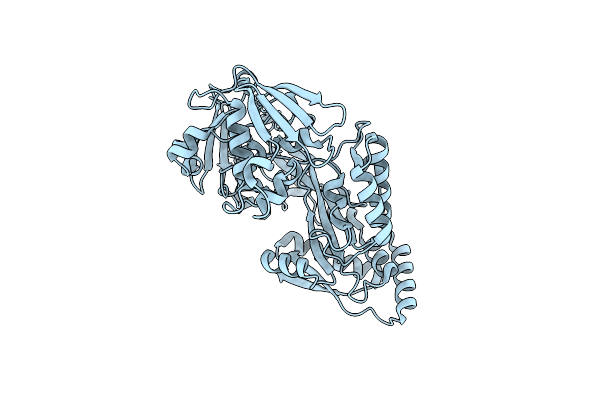
Crystal Structure Of The Peptide Binding Protein Dppe From Bacillus Subtilis In Complex With Murein Tripeptide
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN Ligands: MHI, MG, EDO |
 |
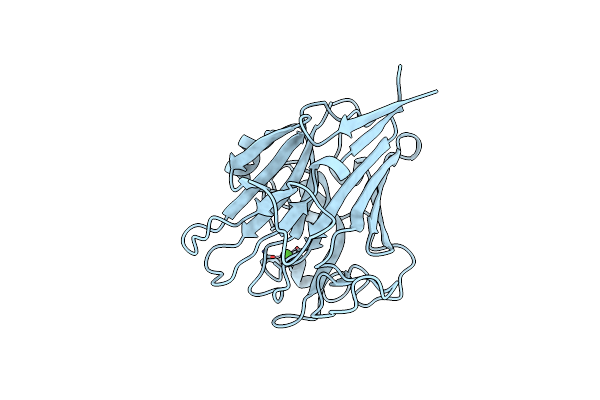
Crystal Structure Of The Peptide Binding Protein Dppe From Bacillus Subtilis In The Unliganded State
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN |
 |
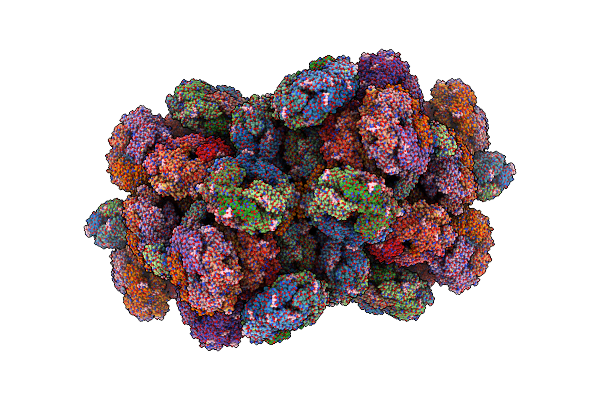
Structure Of Pls A-Domain (Residues 391-656; 513-518 Deletion Mutant) From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-11-09 Classification: CELL ADHESION Ligands: CA |
 |
Structure Of The Phycobilisome From The Red Alga Porphyridium Purpureum In Middle Light
Organism: Porphyridium purpureum
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: PHOTOSYNTHESIS Ligands: PEB, PUB, CYC, PMS |
 |
Organism: Escherichia coli (strain k12), Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2021-11-17 Classification: PEPTIDE BINDING PROTEIN Ligands: GOL |
 |
Crystal Structure Of The Chitinase Domain Of The Spore Coat Protein Cote From Clostridium Difficile
Organism: Peptoclostridium difficile (strain 630), Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-07-22 Classification: STRUCTURAL PROTEIN Ligands: 1PE, PEG |

