Search Count: 37
 |
The Structure Of E. Coli Penicillin Binding Protein 3 (Pbp3) In Complex With A Bicyclic Peptide Inhibitor
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-04-03 Classification: HYDROLASE Ligands: 29N |
 |
Lipidic Cubic Phase Serial Femtosecond Crystallography Structure Of A Photosynthetic Reaction Centre
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, HTO, OLC, SO4, BCB, BPB, UQ1, LDA, FE2, MQ7, NS5 |
 |
Lipidic Cubic Phase Serial Femtosecond Crystallography Structure Of A Photosynthetic Reaction Centre
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, LDA, SO4, BCB, BPB, HTO, UQ2, FE2, MQ9, NS5, OLC |
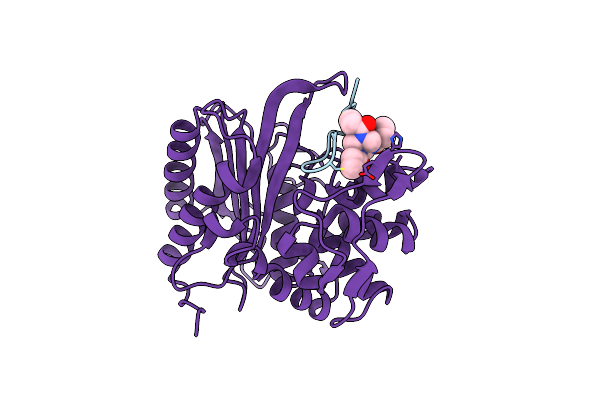
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2021-02-03 Classification: SIGNALING PROTEIN Ligands: ACT, EDO, SO4, NA, 29N |
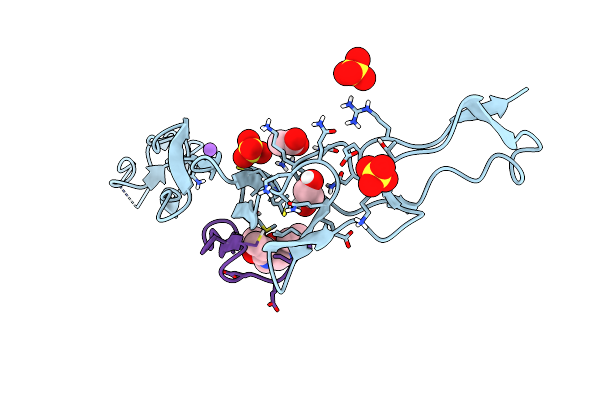
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 1 Ps Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
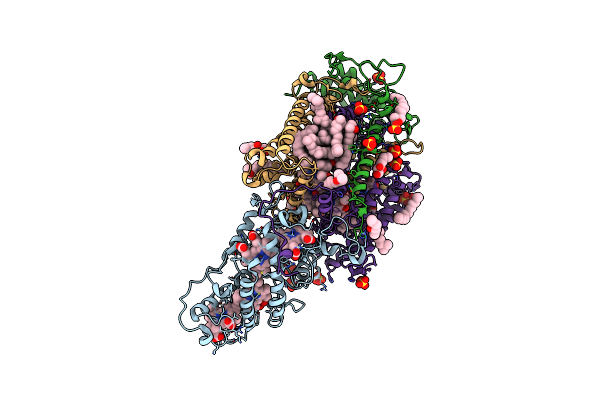
 |
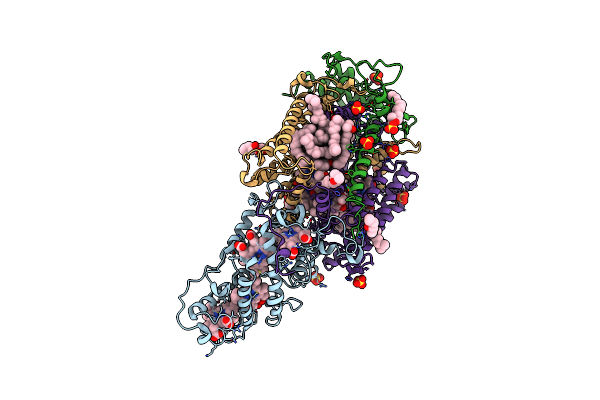
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 5 Ps (A) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
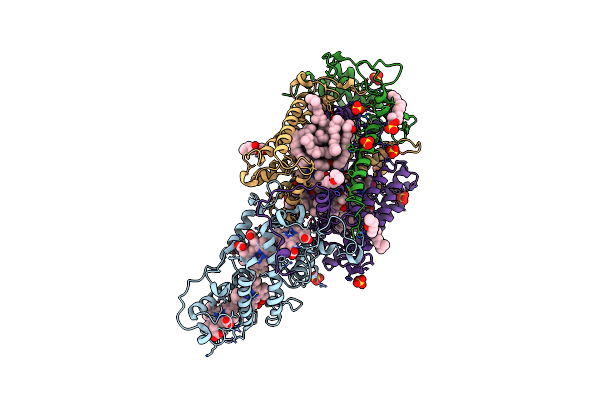
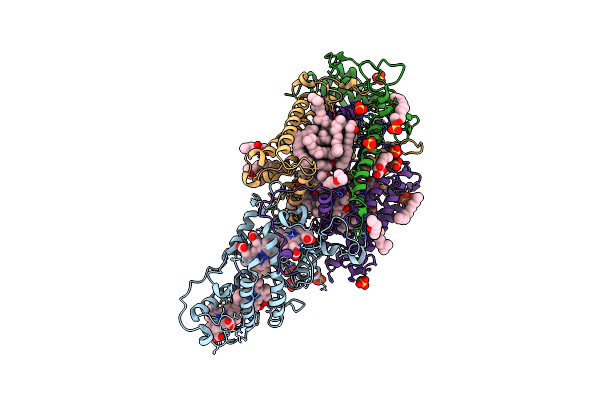
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 300 Ps (A) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
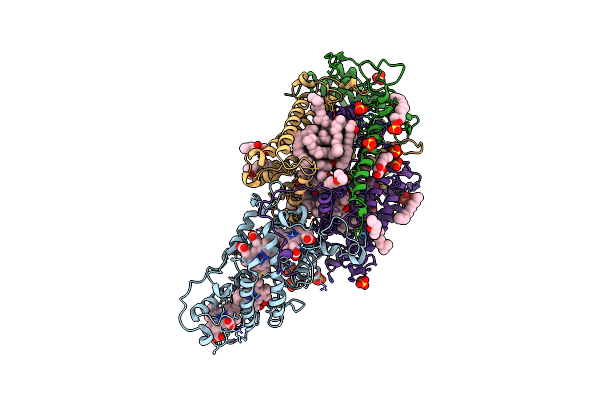
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 20 Ps Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
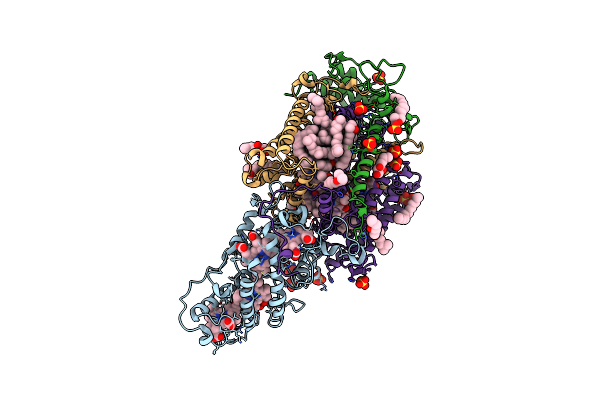
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 300 Ps (B) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 8 Us Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 5 Ps (B) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
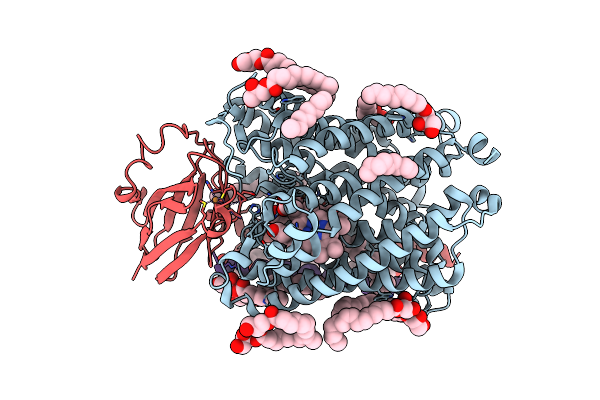
Organism: Human respiratory syncytial virus a (strain a2)
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2018-11-14 Classification: VIRAL PROTEIN Ligands: ZN |
 |
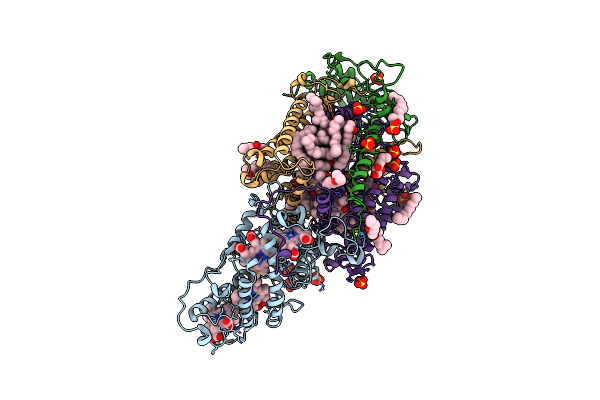
Structure Of Ba3-Type Cytochrome C Oxidase From Thermus Thermophilus By Serial Femtosecond Crystallography
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-08-23 Classification: OXIDOREDUCTASE Ligands: CU, HEM, HAS, OLC, CUA |
 |
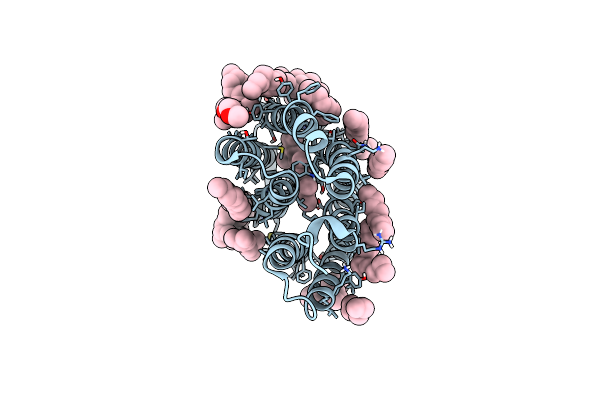
From Macrocrystals To Microcrystals: A Strategy For Membrane Protein Serial Crystallography
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-08-16 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE2, MQ7, NS5 |
 |
From Macrocrystals To Microcrystals: A Strategy For Membrane Protein Serial Crystallography
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-08-16 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE2, MQ7, NS5 |
 |
From Macrocrystals To Microcrystals: A Strategy For Membrane Protein Serial Crystallography
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2017-08-16 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, SO4, FME, LDA, HTO, BCB, BPB, FE2, MQ7, NS5 |
 |
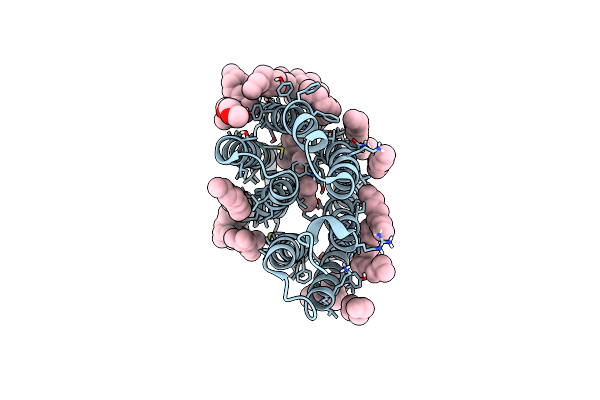
A Three Dimensional Movie Of Structural Changes In Bacteriorhodopsin: Resting State Structure
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-12-21 Classification: PROTON TRANSPORT Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |
 |
A Three Dimensional Movie Of Structural Changes In Bacteriorhodopsin: Structure Obtained 16 Ns After Photoexcitation
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-12-21 Classification: PROTON TRANSPORT Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |
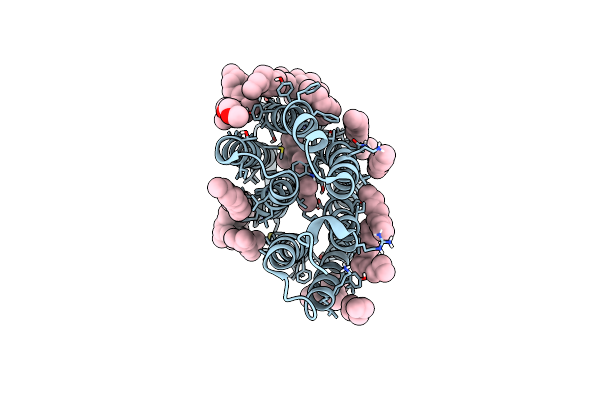 |
A Three Dimensional Movie Of Structural Changes In Bacteriorhodopsin: Structure Obtained 760 Ns After Photoexcitation
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-12-21 Classification: PROTON TRANSPORT Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |
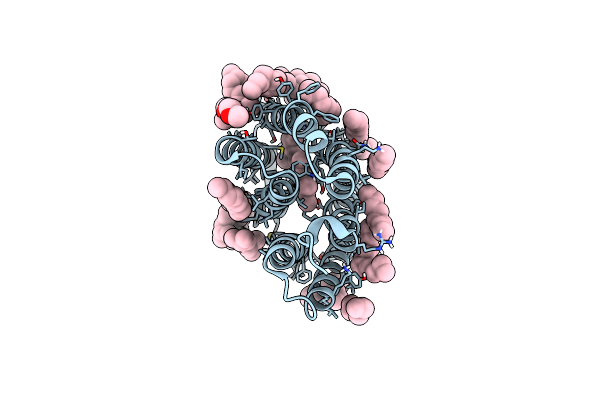 |
A Three Dimensional Movie Of Structural Changes In Bacteriorhodopsin: Structure Obtained 36.2 Us After Photoexcitation
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-12-21 Classification: PROTON TRANSPORT Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |

