Search Count: 49
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: ZN, CA, OQU, EDO, EPE |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: ZN, CA, ORL, EPE, 1PE, EDO |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: ZN, CA, EDO, OQK, EPE |
 |
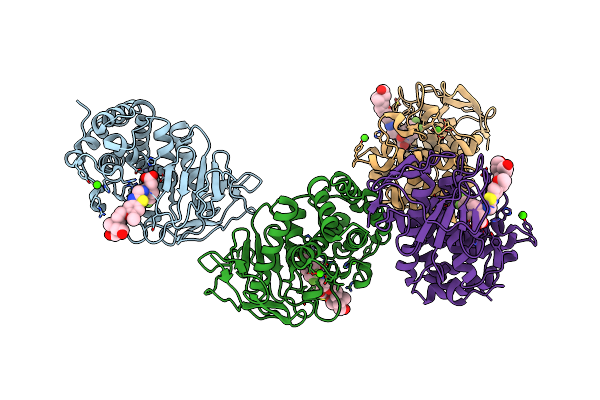
Crystal Structure Of The Vim-2 Acquired Metallo-Beta-Lactamase In Complex With Compound 8 (Jmv-7061)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-04-26 Classification: HYDROLASE Ligands: ZN, ACT, L2R |
 |
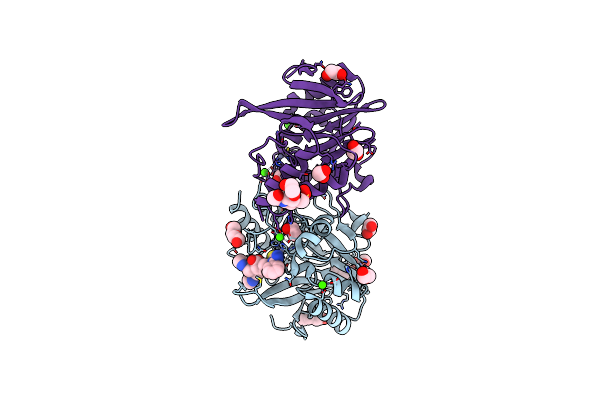
Discovery And Development Of A Novel Inhaled Antivirulence Therapy For The Treatment Of Pseudomonas Aeruginosa Infections In Patients With Chronic Respiratory Disease
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: ZN, CI8, CA |
 |
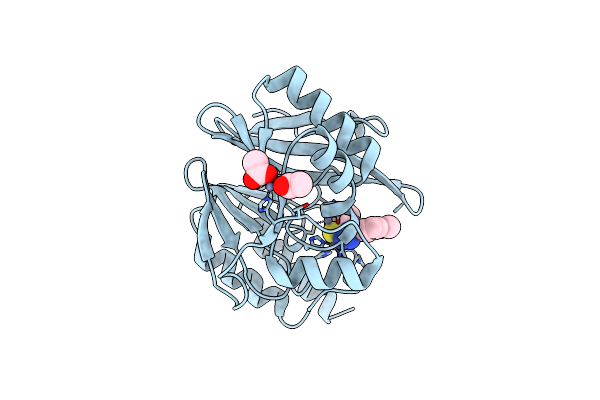
Metallo-Beta-Lactamase Ndm-1 In Complex With 1,2,4-Triazole-3-Thione Compound 26
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-12-14 Classification: HYDROLASE Ligands: ZN, CA, L82, EDO, PEG, EPE |
 |
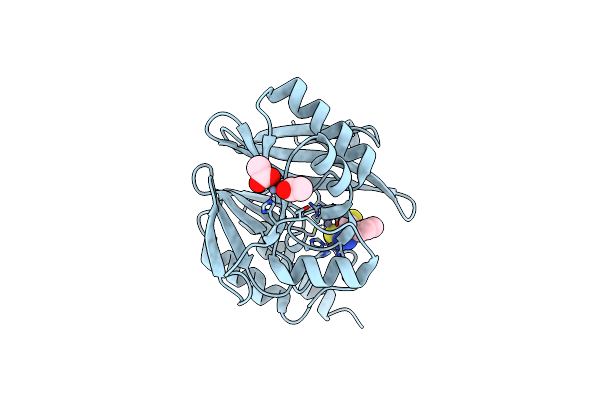
Crystal Structure Of The Vim-2 Acquired Metallo-Beta-Lactamase In Complex With Compound 28 (Jmv-7038)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: ZN, ACT, 7ZN |
 |
Crystal Structure Of The Vim-2 Acquired Metallo-Beta-Lactamase In Complex With Compound 10 (Jmv-7210)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2021-10-20 Classification: HYDROLASE Ligands: ZN, ACT, UNL |
 |
Crystal Structure Of The Vim-2 Acquired Metallo-Beta-Lactamase In Complex With Compound 8 (Jmv-7207)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-10-20 Classification: HYDROLASE Ligands: ZN, ACT, 1TH |
 |
Crystal Structure Of The Vim-2 Acquired Metallo-Beta-Lactamase In Complex With Compound 14 (Jmv-6931)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-10-20 Classification: HYDROLASE Ligands: ZN, ACT, UNL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-08-11 Classification: HYDROLASE Ligands: EDO, 8YL, SO4, CL |
 |
Crystal Structure Of The Oxa-48 Carbapenem-Hydrolyzing Class D Beta-Lactamase In Complex With The Dbo Inhibitor Ant3310
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-08-11 Classification: HYDROLASE Ligands: QS8, EDO, CO2, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-07-21 Classification: HYDROLASE Ligands: QP2, EDO, SO4, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2021-07-21 Classification: HYDROLASE Ligands: 2RG, EDO |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-07-21 Classification: HYDROLASE Ligands: EDO, DWZ |
 |
Crystal Structure Of Class D Beta-Lactamase Oxa-48 In Complex With Ertapenem
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-07-21 Classification: HYDROLASE Ligands: 2RG, EDO, CL, PGE |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: ZN, FMT, N8Z |
 |
Crystal Structure Of The Vim-2 Acquired Metallo-Beta-Lactamase In Complex With Jmv-4690 (Cpd 31)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: ZN, PJB, EDO, ACT, DMS |
 |
Crystal Structure Of The Vim-2 Acquired Metallo-Beta-Lactamase In Complex With The Thiazolecarboxylate Inhibitor Ant2681
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-09-02 Classification: HYDROLASE Ligands: ZN, ACT, QKK |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2020-08-26 Classification: HYDROLASE Ligands: ZN, CA, N8Q |

