Search Count: 6
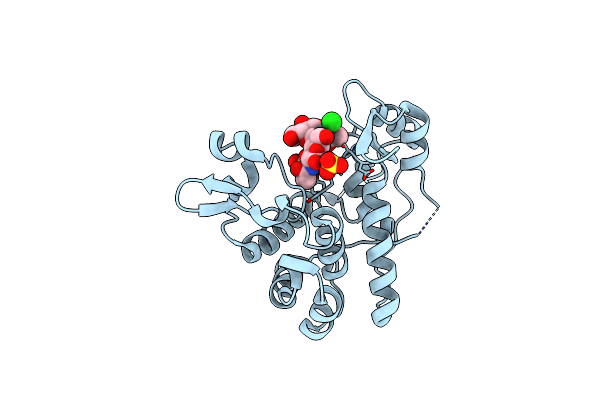 |
Crystal Structure Of S. Aureus Autolysin E In Complex With Disaccharide Nam-Nag
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-10-14 Classification: HYDROLASE Ligands: CL, SO4 |
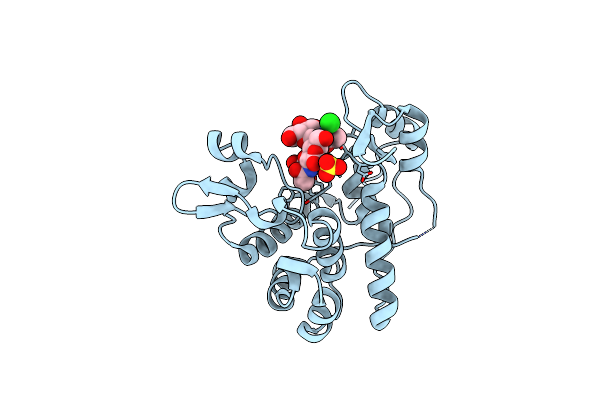 |
Crystal Structure Of Catalytic Mutant E138A Of S. Aureus Autolysin E In Complex With Disaccharide Nag-Nam
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2015-10-14 Classification: HYDROLASE Ligands: CL, SO4 |
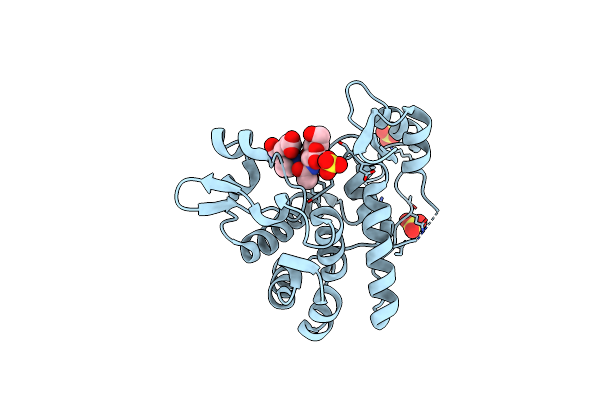 |
Crystal Structure Of S. Aureus Autolysin E In Complex With Muropeptide Nam-L-Ala-D-Iglu
Organism: Staphylococcus aureus (strain mu50 / atcc 700699)
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2015-10-14 Classification: HYDROLASE Ligands: SO4, CL, 3LT |
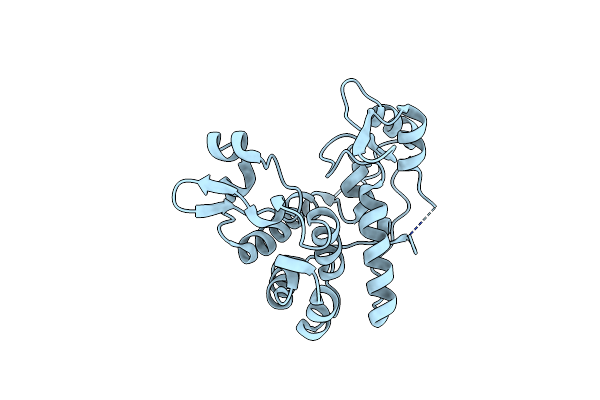 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2015-10-14 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of Stefin A In Complex With Cathepsin H: N-Terminal Residues Of Inhibitors Can Adapt To The Active Sites Of Endo-And Exopeptidases
Organism: Homo sapiens, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-02-18 Classification: HYDROLASE |
 |
Organism: Homo sapiens, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2003-02-18 Classification: HYDROLASE/HYDROLASE INHIBITOR |

