Search Count: 15
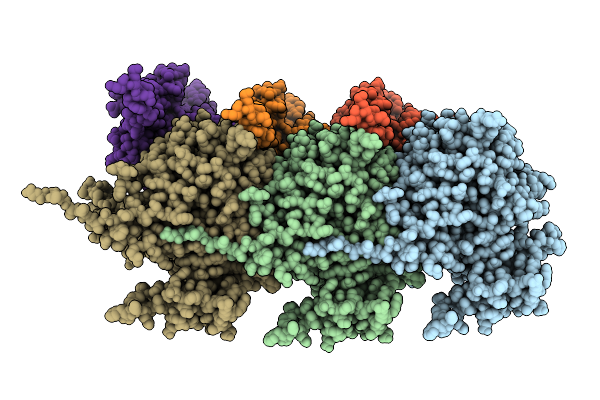 |
Organism: Porphyromonas gingivalis atcc 33277
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN |
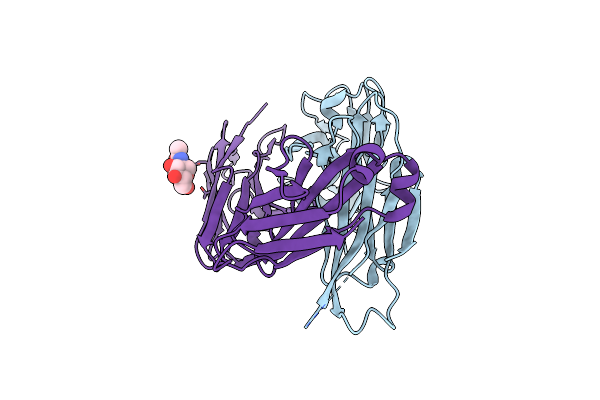 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-10-18 Classification: IMMUNE SYSTEM Ligands: NAG |
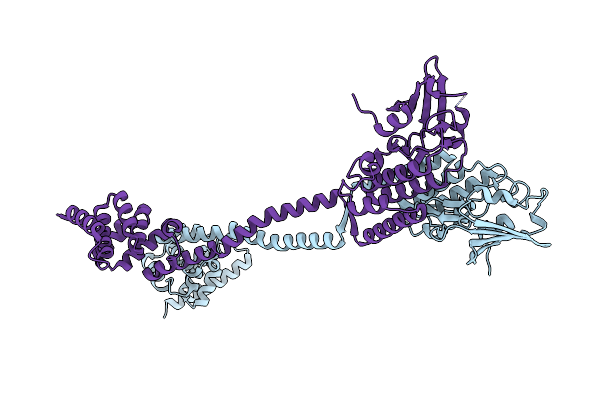 |
Crystal Structure Of The Membrane Pseudokinase Yukc/Essb From Bacillus Subtilis T7Ss
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-05-19 Classification: MEMBRANE PROTEIN |
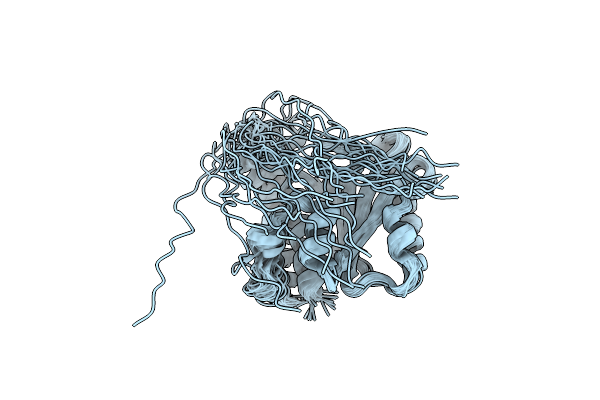 |
Organism: Bacillus subtilis
Method: SOLUTION NMR Release Date: 2020-01-08 Classification: MEMBRANE PROTEIN |
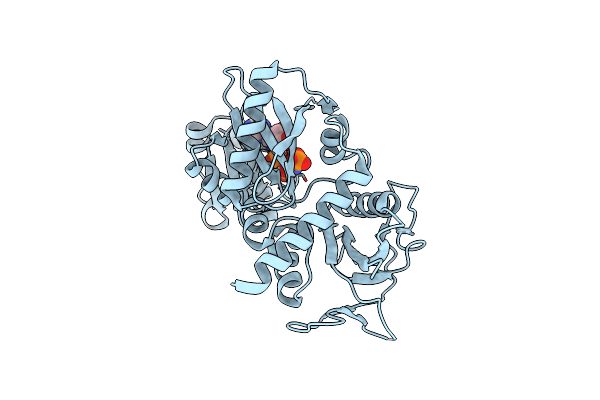 |
Crystal Structure Of D-Alanine-D-Alnine Ligase From Xanthomonas Oryzae Pv. Oryzae With Amppnp
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-02-19 Classification: LIGASE Ligands: ANP, MG |
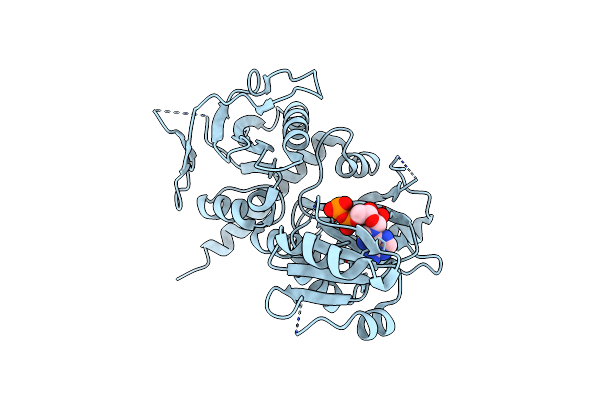 |
Crystal Structure Of D-Alanine-D-Alanine Ligase A From Xanthomonas Oryzae Pathovar Oryzae With Adp
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-02-19 Classification: LIGASE Ligands: MG, ADP |
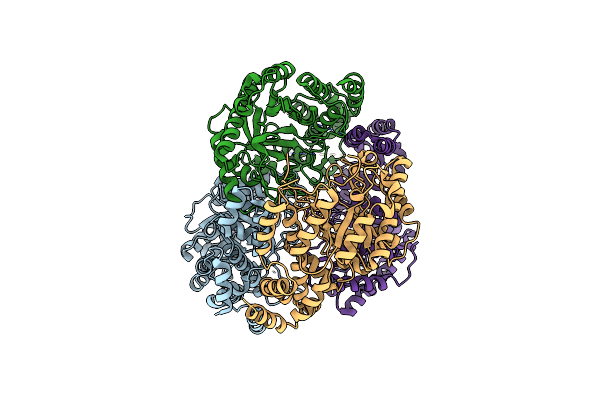 |
Crystal Structure Of L-Rhamnose Isomerase W38A Mutant From Bacillus Halodurans
Organism: Bacillus halodurans
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2012-12-26 Classification: ISOMERASE Ligands: MN |
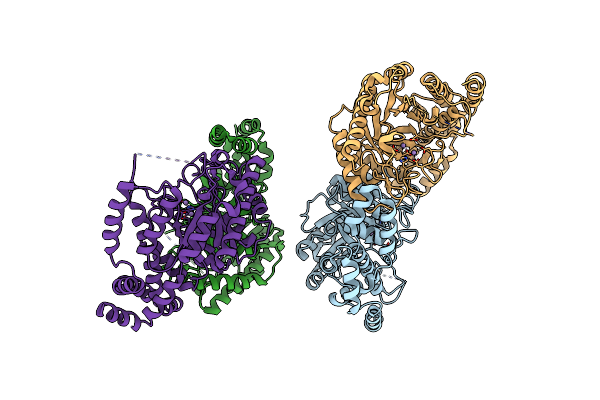 |
Crystal Structure Of L-Rhamnose Isomerase Mutant W38F From Bacillus Halodurans In Complex With Mn
Organism: Bacillus halodurans
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2012-12-05 Classification: ISOMERASE Ligands: MN |
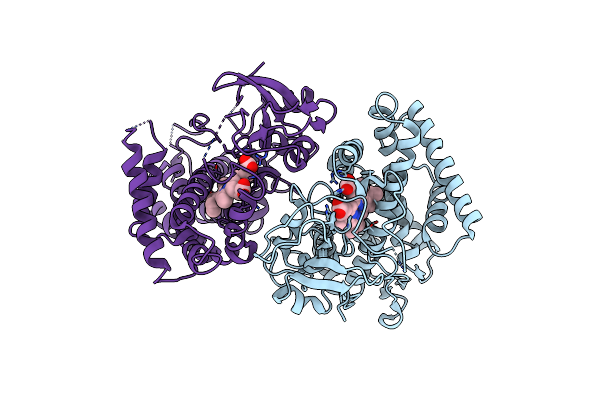 |
Crystal Structure Of Cyp105N1 From Streptomyces Coelicolor: A Cytochrome P450 Oxidase In The Coelibactin Siderophore Biosynthetic Pathway
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2012-11-14 Classification: OXIDOREDUCTASE Ligands: HEM |
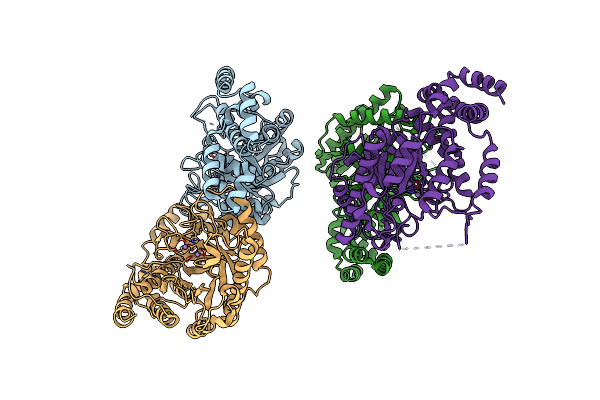 |
Crystal Structure Of L-Rhamnose Isomerase From Bacillus Halodurans In Complex With Mn
Organism: Bacillus halodurans
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-02-01 Classification: ISOMERASE Ligands: MN |
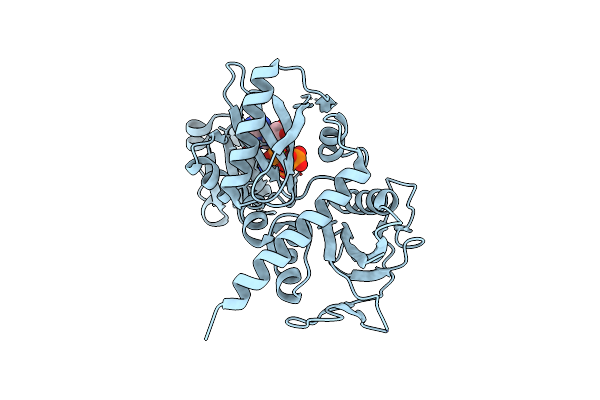 |
Crystal Structure Of D-Alanine-D-Alnine Ligase From Xanthomonas Oryzae Pv. Oryzae With Atp
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2011-05-25 Classification: LIGASE Ligands: ATP, MG |
 |
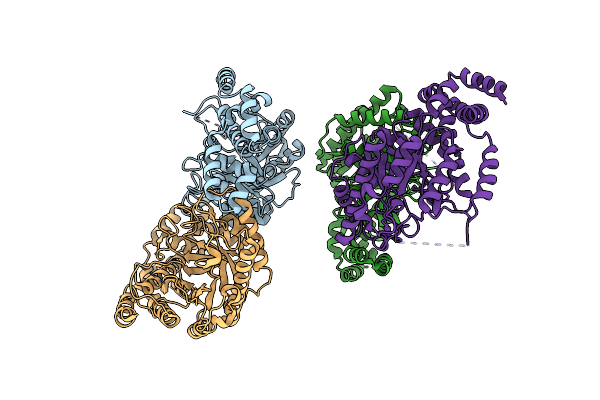
Crystal Structure Of D-Alanine-D-Alanine Ligase A From Xanthomonas Oryzae Pathovar Oryzae With Adp
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-05-04 Classification: LIGASE Ligands: ADP, MG |
 |
Crystal Structure Of L-Rhamnose Isomerase With A Novel High Thermo-Stability From Bacillus Halodurans
Organism: Bacillus halodurans
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2010-11-24 Classification: ISOMERASE |
 |
Crystal Structure Of Co-Chaperonin, Groes (Xoo4289) From Xanthomonas Oryzae Pv. Oryzae Kacc10331
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2010-07-28 Classification: CHAPERONE |
 |
Crystal Structure Of D-Alanine-D-Alanine Ligase From Xanthomonas Oryzae Pv. Oryzae Kacc10331
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-08-18 Classification: LIGASE |

