Search Count: 195
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2024-12-11 Classification: OXIDOREDUCTASE Ligands: A1BH7 |
 |
E.Coli Dsba In Complex With 4-Phenyl-2-(3-Phenylpropyl)Thiazole-5-Carboxylic Acid
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-04-12 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: QVP, CU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-01-18 Classification: TRANSCRIPTION Ligands: WNQ, UNX, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2023-01-18 Classification: TRANSCRIPTION Ligands: X9N, UNX |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: CU, 010 |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: IMD |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: BYZ, PGE |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: PKN, CU |
 |
Crystal Structure Of Ecdsba In A Complex With (1-Methyl-1H-Pyrazol-5-Yl)Methanamine
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: Q2I, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: Q2O, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: Q3F, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: URE, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: DMS, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: GOL, DMS, CU |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-12-29 Classification: OXIDOREDUCTASE Ligands: PEG, TCH |
 |
Crystal Structure Of Vibrio Cholerae Dsba In Complex With Bile Salt Taurocholate
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-12-29 Classification: OXIDOREDUCTASE Ligands: GOL, TCH |
 |
Crystal Structure Of Ribonucleotide Reductase R2 Subunit Solved By Serial Synchrotron Crystallography
Organism: Saccharopolyspora erythraea (strain atcc 11635 / dsm 40517 / jcm 4748 / nbrc 13426 / ncimb 8594 / nrrl 2338)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-10-07 Classification: OXIDOREDUCTASE Ligands: MN3, FE |
 |
Structure Of Galectin-3C In Complex With Lactose Determined By Serial Crystallography Using A Silicon Nitride Membrane Support
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-07-29 Classification: SUGAR BINDING PROTEIN |
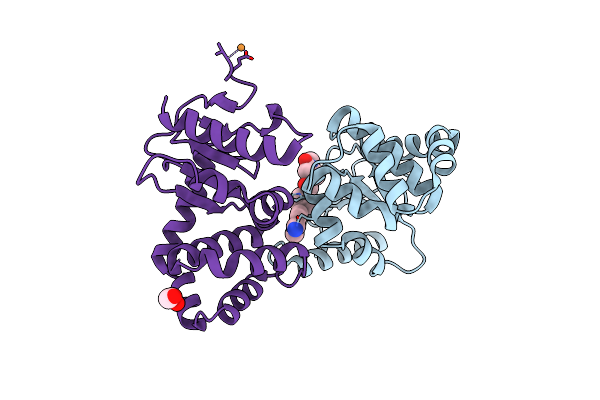 |
Crystal Structure Of Ecdsba In A Complex With Unpurified Reaction Product F1 (Methylpiperazinone 6)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2020-07-01 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: O7P, CU, EDO |
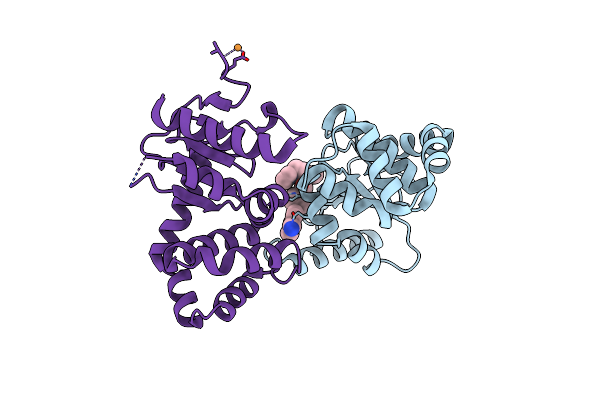 |
Crystal Structure Of Ecdsba In A Complex With Unpurified Reaction Product G6 (Pyrazole 9)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-07-01 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: OAV, CU |

