Search Count: 761
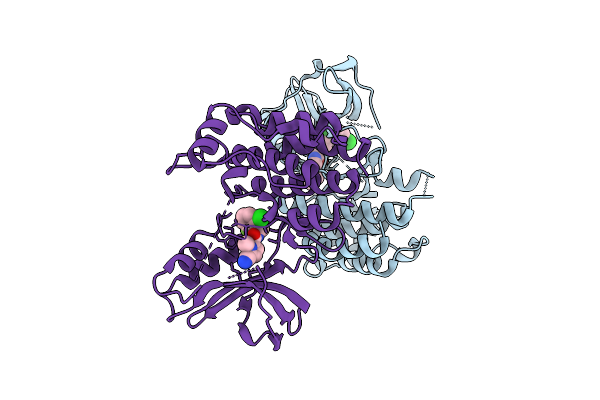 |
Crystal Structure Of An Allosteric Inhibitor Bound To Human Ripk1 Kinase Domain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: A1IX7, IOD, 1HX |
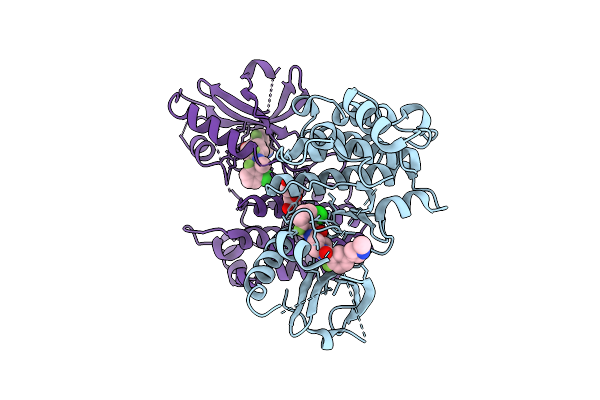 |
Crystal Structure Of An Allosteric Inhibitor Bound To Human Ripk1 Kinase Domain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: A1IX9, IOD, PEG |
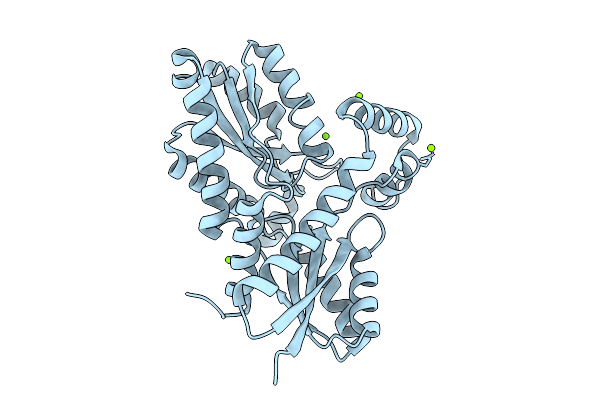 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: MG, CL |
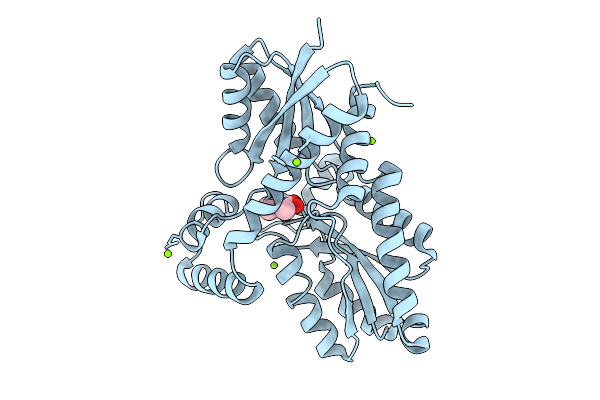 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: 9UM, MG, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: 9UM, MG, MN, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: MG, CL, 9UM, FE2 |
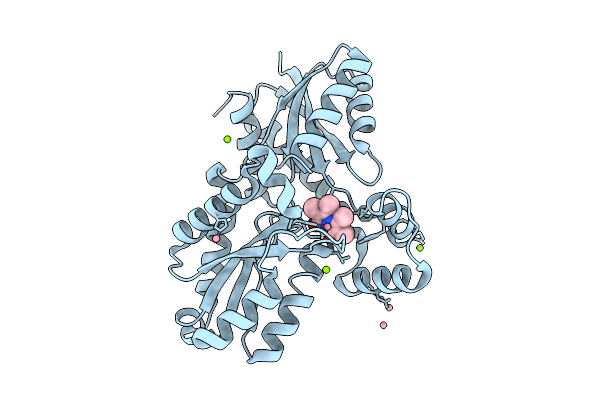 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: CO, MG, CL, 9UM |
 |
Ni(Ii)-Bound Bacillus Subtilis Cpfc (Hemh) Y13C Variant Modified With Bromobimane
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: 9UM, MG, NI, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: 9UM, CU, MG, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, MG, 9UM |
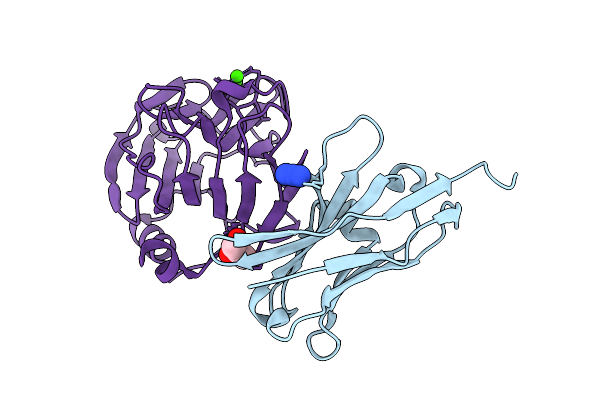 |
Crystal Structure Of The Complex Between Neuronal Pentraxin 2 (Np2 Ptx) And Antibody Fragment Vhh N1
Organism: Vicugna pacos, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: PROTEIN BINDING Ligands: AZI, CA, CL, GOL |
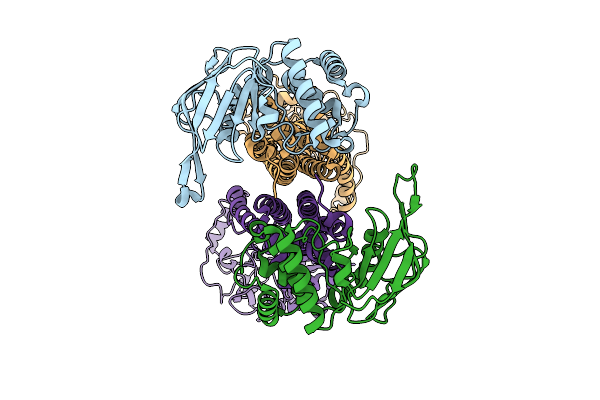 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN |
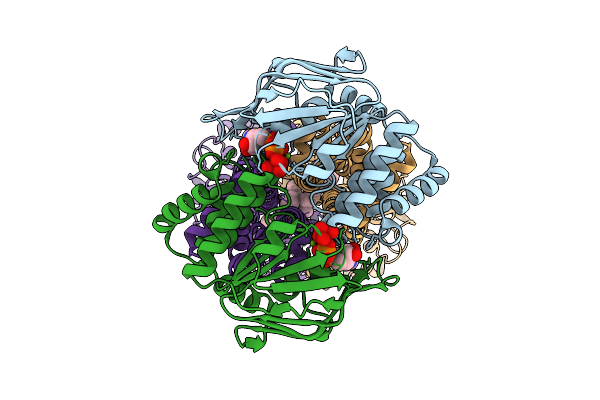 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: ADP, VO4, MG, UNL |
 |
Organism: Vicugna pacos
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: IMMUNE SYSTEM Ligands: SO4, CL, GOL, 1PE |
 |
Organism: Vicugna pacos
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: IMMUNE SYSTEM Ligands: SO4, NA, GOL |
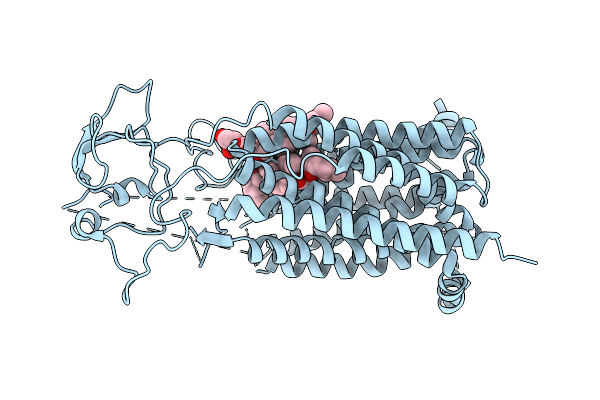 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: TRANSPORT PROTEIN Ligands: AJP |
 |
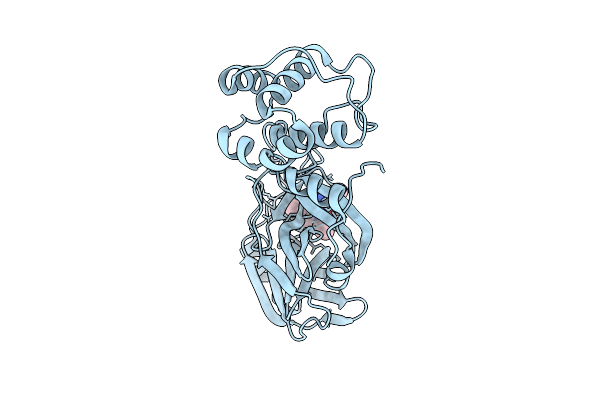
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro)In Complex With Inhibitor Avi-3318
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1BTM |
 |
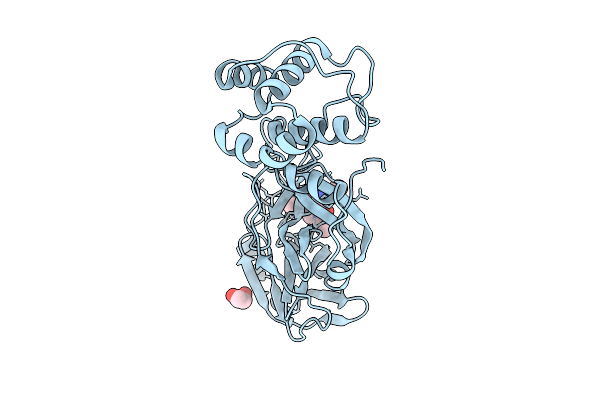
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With Inhibitor Avi-4692
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE Ligands: A1BVT, A1BTO |
 |
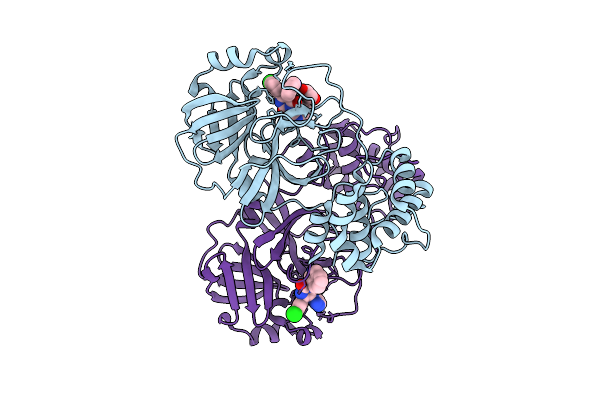
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro)In Complex With Inhibitor Avi-4516
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1BTP, EDO |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) Variant Q192T In Complex With Inhibitor Avi-4303
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: EDO, A1BTN, CL |

