Search Count: 409
 |
Crystal Structure Of Ice Binding Protein From Candidatus Cryosericum Odellii Smc5
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: RECOMBINATION |
 |
Crystal Structure Of Ice Binding Protein From Candidatus Cryosericum Odellii Smc5
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: RECOMBINATION |
 |
Crystal Structure Of An Ice-Binding Protein From Candidatus Cryosericum Odellii
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PROTEIN BINDING |
 |
Crystal Structure Of Engineered Ice-Binding Protein (M74I And A97V) From Candidatus Cryosericum Odellii Strain Smc5_169
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PROTEIN BINDING |
 |
Crystal Structure Of Cyp46A1 With [(1R,5S)-3-Oxa-8-Azabicyclo[3.2.1]Octan-8-Yl][(4R,8M)-8-(1,3-Oxazol-5-Yl)-6-(Trifluoromethyl)Imidazo[1,2-A]Pyridin-3-Yl]Methanone (Compound 3K)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE/INHIBITOR Ligands: HEM, A1BZE |
 |
Crystal Structure Of Cyp46A1 With (Morpholin-4-Yl)[(4R,8M)-8-(1,3-Oxazol-5-Yl)-6-(Trifluoromethyl)Imidazo[1,2-A]Pyridin-3-Yl]Methanone (Compound 2H)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE/INHIBITOR Ligands: HEM, A1BZD |
 |
Crystal Structure Of Cyp46A1 With Cyclopropyl[(4M)-4-(1,3-Oxazol-5-Yl)-6-(Trifluoromethyl)-1H-Indol-1-Yl]Methanone (Compound 2B)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE/INHIBITOR Ligands: HEM, A1BZC, EDO |
 |
Crystal Structure Of Transcriptional Regulator (Nrpr) From Streptococcus Salivarius K12
Organism: Streptococcus salivarius k12
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: TRANSCRIPTION |
 |
Structure Of Rat Neuronal Nitric Oxide Synthase R349A Mutant Heme Domain Bound With 4-Methyl-6-(3-((Methylamino)Methyl)Phenyl)Pyridin-2-Amine Dihydrochloride
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, KMI, ACT, ZN |
 |
Structure Of Rat Nnos R349A Mutant Heme Domain Bound With 4-Methyl-6-(3-((Methylamino)Methyl)-5-(Trifluoromethyl)Phenyl)Pyridin-2-Amine Dihydrochloride
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, ACT, ZN, A1A0F |
 |
Structure Of Rat Neuronal Nitric Oxide Synthase R349A Mutant Heme Domain Bound With 6-(3-Chloro-5-((Methylamino)Methyl)Phenyl)-4-Methylpyridin-2-Amine Dihydrochloride
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, A1A0G, ACT, ZN |
 |
Structure Of Rat Neuronal Nitric Oxide Synthase R349A Mutant Heme Domain Bound With 6-(3-Chloro-5-((Dimethylamino)Methyl)Phenyl)-4-Methylpyridin-2-Amine Dihydrochloride
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, ACT, ZN, A1A0N |
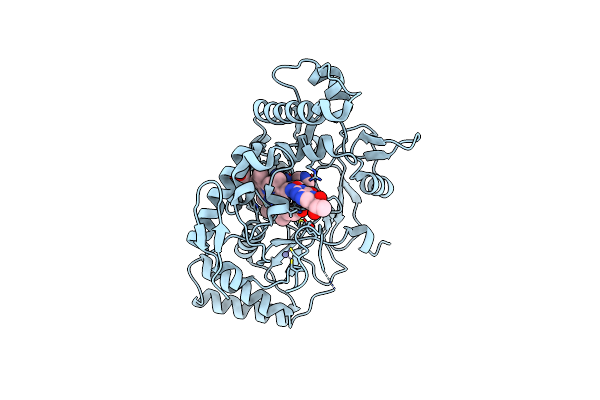 |
Structure Of Rat Neuronal Nitric Oxide Synthase R349A Mutant Bound With 4-Methyl-5'-((Methylamino)Methyl)-[2,3'-Bipyridin]-6-Amine Dihydrochloride
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, ACT, ZN, A1A0E |
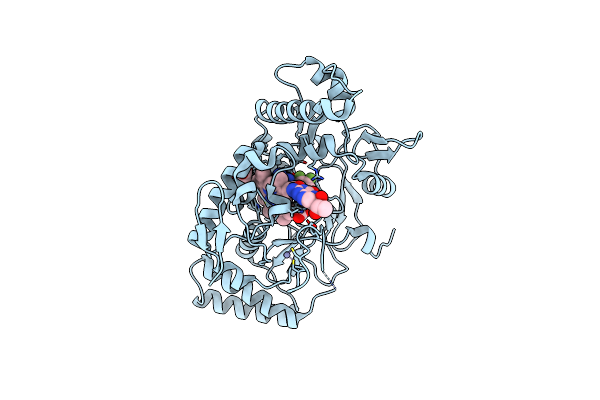 |
Structure Of Rat Neuronal Nitric Oxide Synthase Bound With 6-(2,3-Difluoro-5-((Methylamino)Methyl)Phenyl)-4-Methylpyridin-2-Amine Dihydrochloride
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: HEM, H4B, A1A0B, ACT, ZN |
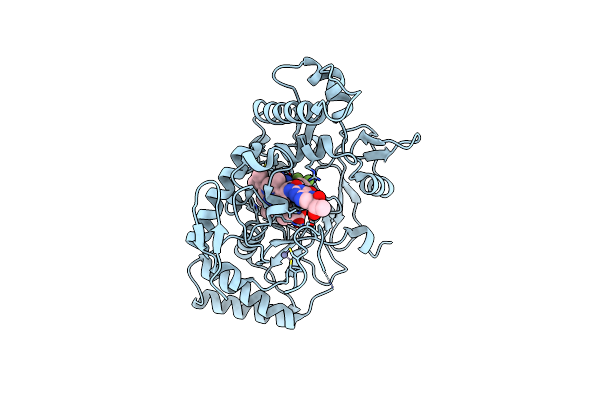 |
Structure Of Rat Neuronal Nitric Oxide Synthase R349A Mutant Heme Domain Bound With 6-(5-((Dimethylamino)Methyl)-2,3-Difluorophenyl)-4-Methylpyridin-2-Amine Dihydrochloride
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, ZN, A1A0K |
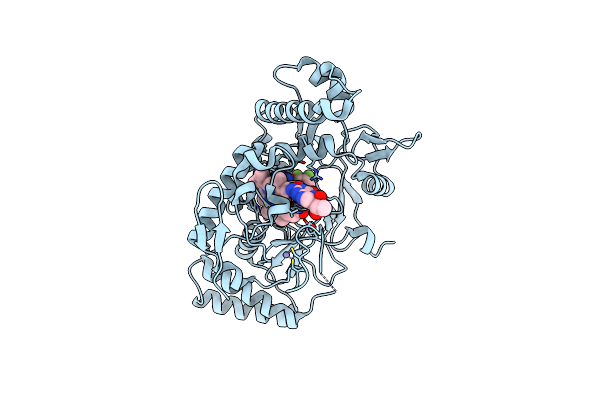 |
Structure Of Rat Neuronal Nitric Oxide Synthase R349A Mutant Bound With 6-(2,3-Difluoro-5-(((2-Fluoroethyl)Amino)Methyl)Phenyl)-4-Methylpyridin-2-Amine Dihydrochloride
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, ZN, A1AZ9 |
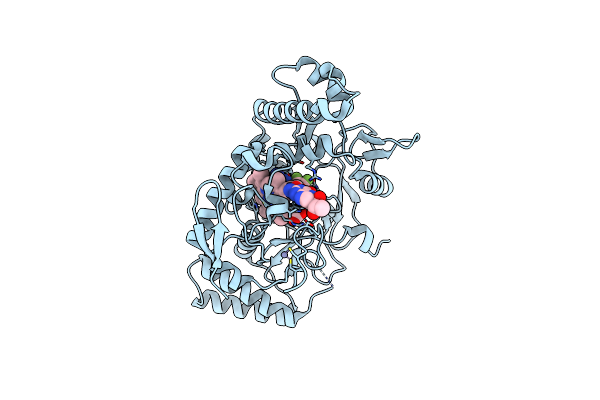 |
Structure Of Rat Neuronal Nitric Oxide Synthase R349A Mutant Heme Domain Bound With 6-(5-((3,3-Difluoroazetidin-1-Yl)Methyl)-2,3-Difluorophenyl)-4-Methylpyridin-2-Amine Dihydrochloride
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, ZN, A1A0D |
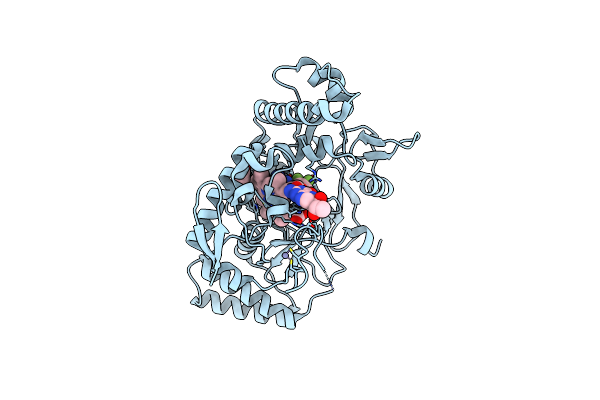 |
Structure Of Rat Neuronal Nitric Oxide Synthase R349A Mutant Heme Domain Bound With 6-(2,3-Difluoro-5-(2-(Methylamino)Ethyl)Phenyl)-4-Methylpyridin-2-Amine Dihydrochloride
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, A1A0C, ACT, ZN |
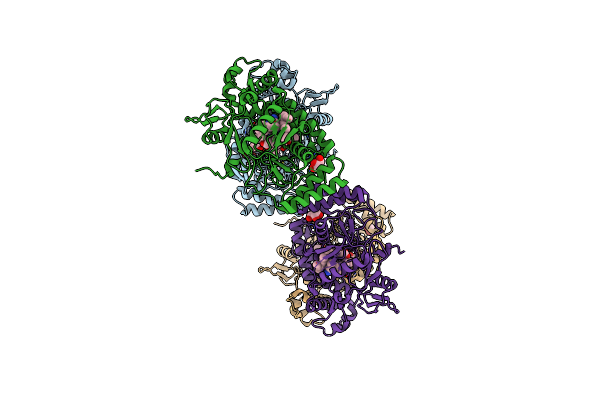 |
Structure Of Human Neuronal Nitric Oxide Synthase R354A/G357D Mutant Heme Domain Bound With 4-Methyl-6-(3-((Methylamino)Methyl)Phenyl)Pyridin-2-Amine Dihydrochloride
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: HEM, H4B, KMI, GOL, ZN |
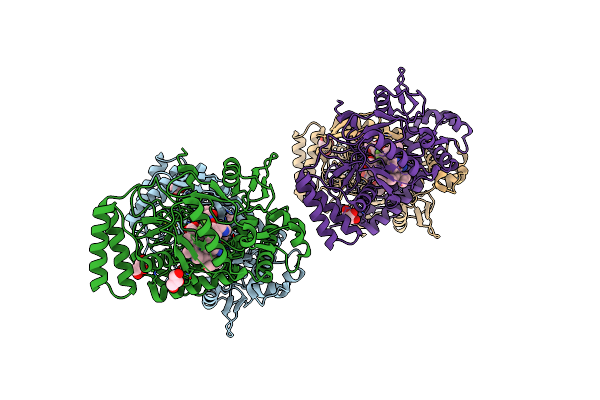 |
Structure Of Human Neuronal Nitric Oxide Synthase R354A/G357D Mutant Heme Domain Bound With 4-Methyl-6-(3-((Methylamino)Methyl)-5-(Trifluoromethyl)Phenyl)Pyridin-2-Amine Dihydrochloride
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: HEM, H4B, A1A0F, GOL, ZN |

