Search Count: 16
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-10-07 Classification: UNKNOWN FUNCTION |
 |
Solution Structure Of Ph1500: A Homohexameric Protein Centered On A 12-Bladed Beta-Propeller
Organism: Pyrococcus horikoshii
Method: SOLUTION NMR Release Date: 2014-01-29 Classification: UNKNOWN FUNCTION |
 |
Structure And Activity Of The N-Terminal Substrate Recognition Domains In Proteasomal Atpases - The Arc Domain Structure
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-05-12 Classification: ATP-BINDING PROTEIN |
 |
Proteasome-Activating Nucleotidase (Pan) N-Domain (57-134) From Archaeoglobus Fulgidus Fused To Gcn4
Organism: Saccharomyces cerevisiae, Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-04-28 Classification: TRANSCRIPTION,HYDROLASE |
 |
Proteasome-Activating Nucleotidase (Pan) N-Domain (57-134) From Archaeoglobus Fulgidus Fused To Gcn4, P61A Mutant
Organism: Saccharomyces cerevisiae, Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-04-28 Classification: TRANSCRIPTION,HYDROLASE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2008-11-18 Classification: TRANSCRIPTION |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-11-18 Classification: TRANSCRIPTION |
 |
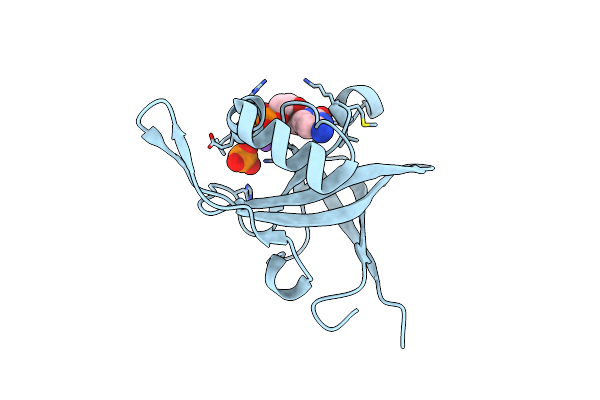
Riboflavin Kinase Mj0056 From Methanocaldococcus Jannaschii In Complex With Po4
Organism: Methanococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2007-11-20 Classification: TRANSFERASE Ligands: PO4, ZN, CL |
 |
Riboflavin Kinase Mj0056 From Methanocaldococcus Jannaschii In Complex With Cdp And Po4
Organism: Methanococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2007-11-20 Classification: TRANSFERASE Ligands: CDP, NA, PO4 |
 |
Riboflavin Kinase Mj0056 From Methanocaldococcus Jannaschii In Complex With Cdp
Organism: Methanococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2007-11-20 Classification: TRANSFERASE Ligands: CDP, MG, MRD |
 |
Riboflavin Kinase Mj0056 From Methanocaldococcus Jannaschii In Complex With Cdp And Fmn
Organism: Methanococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-11-20 Classification: TRANSFERASE Ligands: CDP, MG, IOD, CL, FMN |
 |
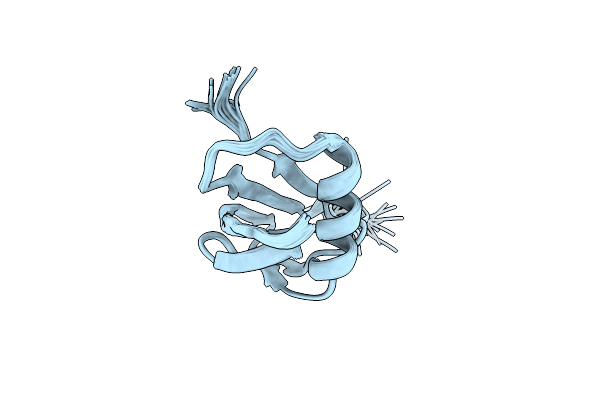
Organism: Methanocaldococcus jannaschii
Method: SOLUTION NMR Release Date: 2007-10-09 Classification: TRANSFERASE |
 |
Organism: Pyrococcus horikoshii
Method: SOLUTION NMR Release Date: 2007-09-25 Classification: UNKNOWN FUNCTION |
 |
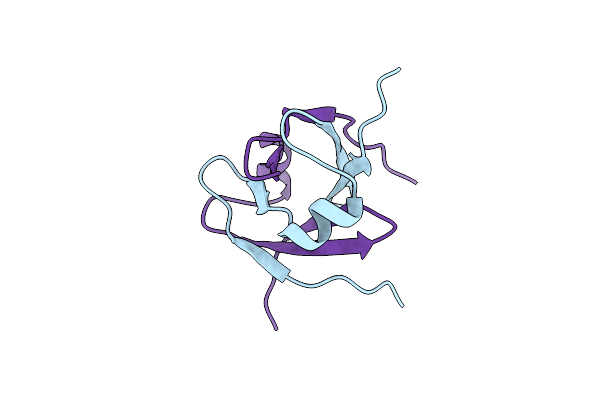
Organism: Pyrococcus horikoshii
Method: SOLUTION NMR Release Date: 2006-12-05 Classification: TRANSCRIPTION |
 |
Organism: Bacillus subtilis
Method: SOLUTION NMR Release Date: 2005-04-12 Classification: TRANSCRIPTION |
 |
Organism: Bacillus subtilis
Method: SOLUTION NMR Release Date: 2005-04-12 Classification: TRANSCRIPTION |

