Search Count: 125
 |
The Full-Length Human Sweet Taste Receptor Tas1R2 And Tas1R3 In The Apo State
Organism: Homo sapiens, Discosoma sp. (sea anemone), Branchiostoma lanceolatum
Method: ELECTRON MICROSCOPY Resolution:3.41 Å Release Date: 2025-07-16 Classification: SIGNALING PROTEIN |
 |
The Vft Domains Of Human Sweet Taste Receptor Tas1R2 And Tas1R3 In The Apo State
Organism: Homo sapiens, Discosoma sp. (sea anemone), Branchiostoma lanceolatum
Method: ELECTRON MICROSCOPY Resolution:3.18 Å Release Date: 2025-07-16 Classification: SIGNALING PROTEIN |
 |
The Transmembrane Domains Of Human Sweet Taste Receptor Tas1R2 And Tas1R3 In The Apo State
Organism: Homo sapiens, Discosoma sp. (sea anemone), Branchiostoma lanceolatum
Method: ELECTRON MICROSCOPY Resolution:3.77 Å Release Date: 2025-07-16 Classification: SIGNALING PROTEIN |
 |
The Full-Length Human Sweet Taste Receptor Tas1R2 And Tas1R3 In The Sucralose-Bound State
Organism: Homo sapiens, Discosoma sp., Branchiostoma lanceolatum
Method: ELECTRON MICROSCOPY Resolution:3.59 Å Release Date: 2025-07-16 Classification: SIGNALING PROTEIN |
 |
The Vft Domains Of Human Sweet Taste Receptor Tas1R2 And Tas1R3 In The Sucralose-Bound State
Organism: Homo sapiens, Discosoma sp., Branchiostoma lanceolatum
Method: ELECTRON MICROSCOPY Resolution:3.33 Å Release Date: 2025-07-16 Classification: SIGNALING PROTEIN |
 |
Organism: Macaca mulatta, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: IMMUNE SYSTEM Ligands: NAG |
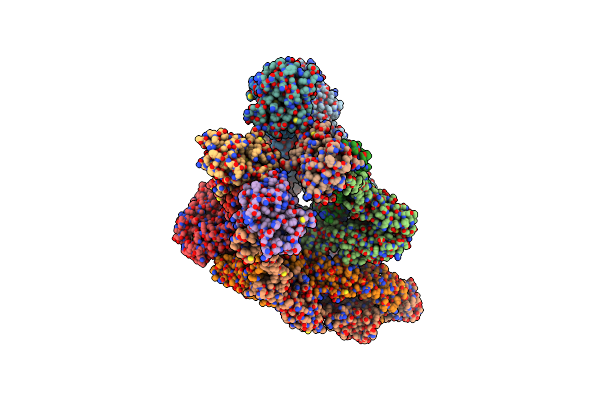 |
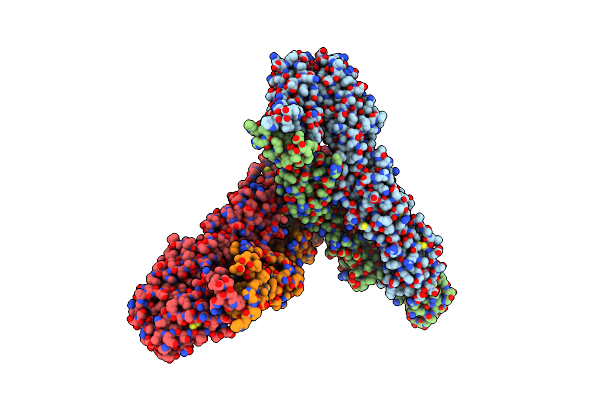
Structure Of Western Equine Encephalitis Virus 71V1658 Strain Vlp In Complex With Human Pcdh10 Ec1
Organism: Western equine encephalitis virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: NAG |
 |
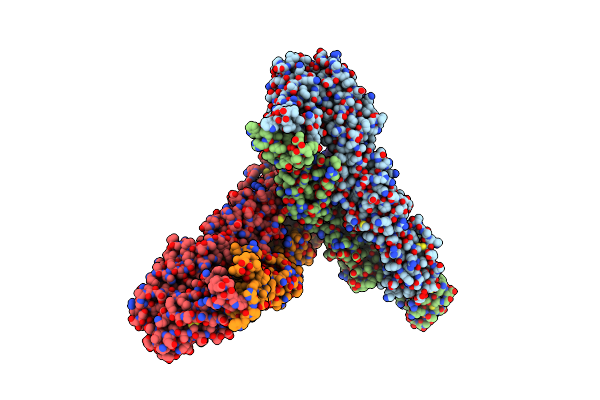
Structure Of Western Equine Encephalitis Virus Mcmillan Strain In Complex With Vldlr La1-2
Organism: Western equine encephalitis virus, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.60 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: CA |
 |
Structure Of Western Equine Encephalitis Virus Mcmillan Strain In Complex With Vldlr La2-3
Organism: Western equine encephalitis virus, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.90 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: CA |
 |
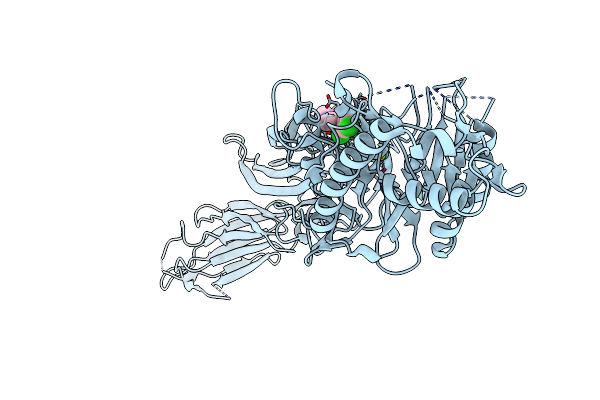
Potent Inhibition Of The Protein Arginine Deiminases (Pad1-4) By Targeting A Ca2+ Dependent Allosteric Binding Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: CA, A1A8O |
 |
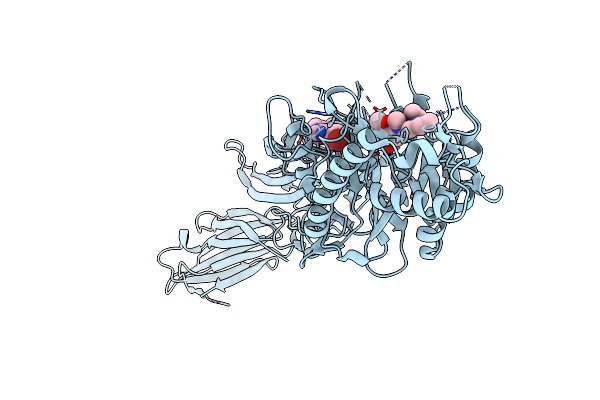
Inhibiting Peptidylarginine Deiminases (Pad1-4) By Targeting A Ca2+ Dependent Allosteric Binding Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2025-05-28 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, A1BZK |
 |
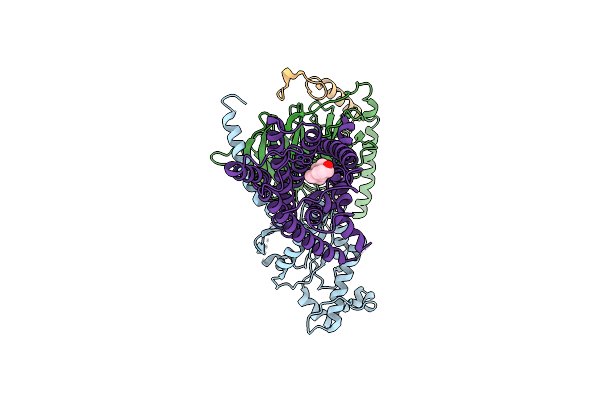
Potent Inhibition Of The Protein Arginine Deiminases (Pad1-4) By Targeting A Ca2+ Dependent Allosteric Binding Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2025-05-28 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1A9P, 3YZ |
 |
Identification, Structure And Agonist Design Of An Androgen Membrane Receptor.
Organism: Rattus norvegicus, Bos taurus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: MEMBRANE PROTEIN Ligands: DHT |
 |
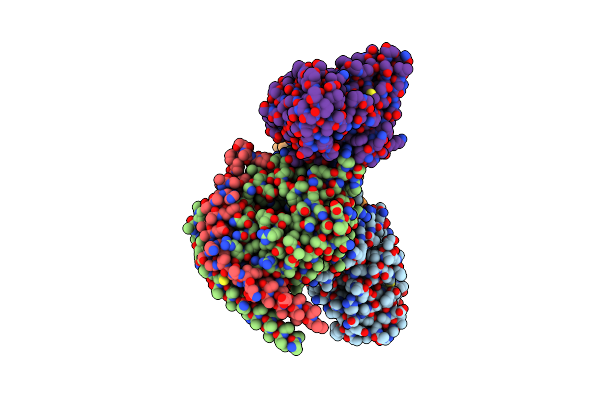
Identification, Structure And Agonist Design Of An Androgen Membrane Receptor
Organism: Bos taurus, Rattus norvegicus, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.75 Å Release Date: 2025-02-12 Classification: MEMBRANE PROTEIN Ligands: YNH |
 |
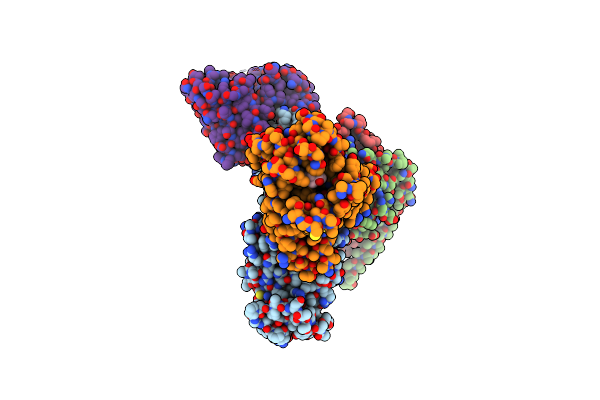
Identification, Structure And Agonist Design Of An Androgen Membrane Receptor
Organism: Rattus norvegicus, Bos taurus, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.88 Å Release Date: 2025-02-12 Classification: MEMBRANE PROTEIN Ligands: A1LU1 |
 |
Identification, Structure And Agonist Design Of An Androgen Membrane Receptor.
Organism: Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: MEMBRANE PROTEIN Ligands: DHT |
 |
Identification, Structure And Agonist Design Of An Androgen Membrane Receptor.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: MEMBRANE PROTEIN Ligands: DHT |
 |
Organism: Candidatus accumulibacter adiacens, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Candidatus accumulibacter adiacens, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
Unveiling The Role Of Human Prohibitin 2 As A Mitochondrial Calcium Channel In Parkinson'S Disease
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2024-06-05 Classification: TRANSPORT PROTEIN |

