Search Count: 22
 |
Crystal Structure Of The Catalytic Domain Of An Aa9 Lytic Polysaccharide Monooxygenase From Thermothelomyces Thermophilus (Ttlpmo9F)
Organism: Thermothelomyces thermophilus atcc 42464
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2024-12-25 Classification: METAL BINDING PROTEIN Ligands: PEG, PG4, EDO, FMT, CU |
 |
Crystal Structure Of Thermothelomyces Thermophila Gh30 (Double Mutant Ee) In Complex With Xylopentaose
Organism: Thermothelomyces
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-05-22 Classification: HYDROLASE Ligands: EDO, F |
 |
Crystal Structure Of Thermothelomyces Thermophila Gh30 (Double Mutant Ee) In Complex With Xylotriose.
Organism: Thermothelomyces
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-05-22 Classification: HYDROLASE Ligands: EDO, PEG |
 |
Crystal Structure Of Thermothelomyces Thermophila (Double Mutant Ee) In Complex With Aldotetrauronic Acid
Organism: Thermothelomyces thermophilus
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2024-05-22 Classification: HYDROLASE Ligands: EDO |
 |
Organism: Thermothelomyces thermophilus
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2024-03-20 Classification: HYDROLASE Ligands: B3P, PEG, ACT, EDO, NAG, PG4 |
 |
Organism: Thermothelomyces thermophilus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-08-23 Classification: HYDROLASE Ligands: NAG, ACT, K, B3P, SCN, EDO |
 |
The Crystal Structure Of A Feruloyl Esterase C From Fusarium Oxysporum In Complex With P-Coumaric Acid
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2023-07-05 Classification: HYDROLASE Ligands: HC4, NAG, EDO, CA, PG4, P6G, PEG |
 |
Crystal Structure Of Laccase-Like Multicopper Oxidase (Lmco) From Thermothelomyces Thermophilus
Organism: Thermothelomyces thermophilus atcc 42464
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-06-28 Classification: OXIDOREDUCTASE Ligands: NAG, EDO, GOL, OXY, CU |
 |
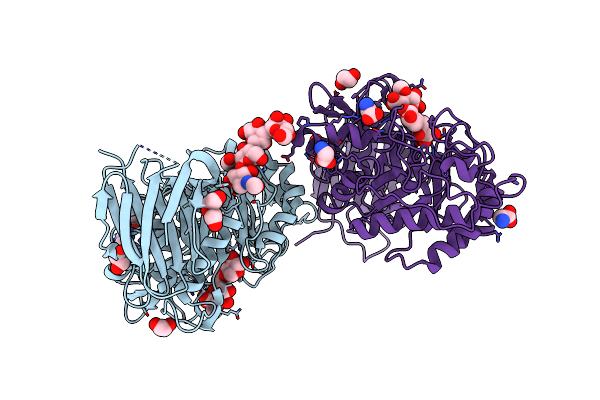
Crystal Structure Of Gh30 (Double Mutant Ee) From Thermothelomyces Thermophila.
Organism: Myceliophthora thermophila (strain atcc 42464 / bcrc 31852 / dsm 1799)
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2021-09-08 Classification: HYDROLASE Ligands: GOL, PEG, EDO, NAG |
 |
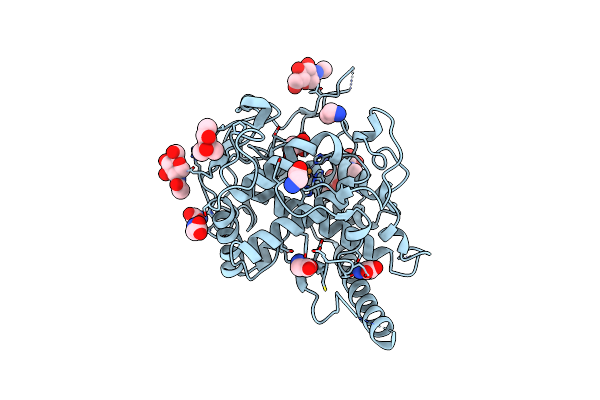
Crystal Structure Of Gh30 (Mutant E188A) Complexed With Aldotriuronic Acid From Thermothelomyces Thermophila.
Organism: Myceliophthora thermophila (strain atcc 42464 / bcrc 31852 / dsm 1799)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-09-08 Classification: HYDROLASE Ligands: GLY, EDO, PEG |
 |
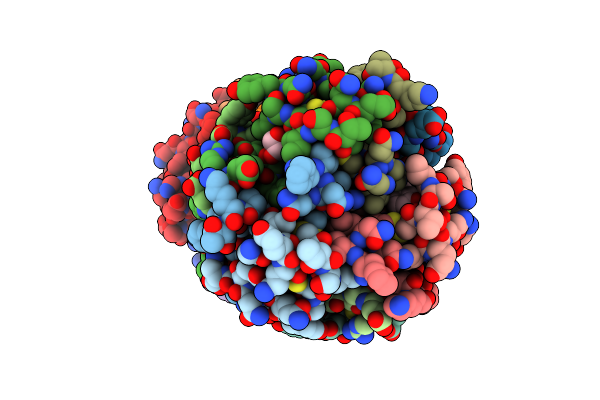
Structure Of Polyphenol Oxidase (Mutant G292N) From Thermothelomyces Thermophila
Organism: Thermothelomyces thermophilus atcc 42464
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2021-03-24 Classification: OXIDOREDUCTASE Ligands: NAG, MPD, GLY, SER, CU |
 |
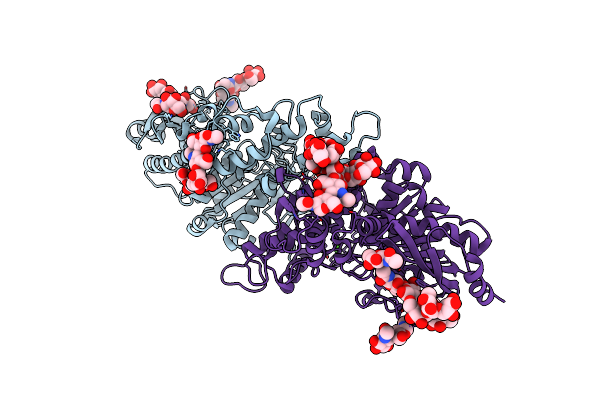
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-06-12 Classification: HORMONE Ligands: CRS, EDO, ZN, IS8 |
 |
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-01-30 Classification: HYDROLASE Ligands: NAG, CA |
 |
Organism: Heterobasidion irregulare tc 32-1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-05-16 Classification: OXIDOREDUCTASE Ligands: NAG, CU, GOL, AKR, NA |
 |
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-12-07 Classification: OXIDOREDUCTASE Ligands: MAN, CU, SO4, LI |
 |
Organism: Fusarium oxysporum f. sp. raphani
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-09-02 Classification: HYDROLASE Ligands: MPD |
 |
The Structure Of The Catalytic Domain Of Nclpmo9C From The Filamentous Fungus Neurospora Crassa
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2015-07-22 Classification: OXIDOREDUCTASE Ligands: CU, GOL |
 |
The Structure Of The Catalytic Domain Of Nclpmo9C From The Filamentous Fungus Neurospora Crassa
Organism: Neurospora crassa (strain atcc 24698 / 74-or23-1a / cbs 708.71 / dsm 1257 / fgsc 987)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-07-22 Classification: OXIDOREDUCTASE Ligands: ZN, ACT, GOL |
 |
Crystal Structure Of Recombinant Glucuronoyl Esterase From Sporotrichum Thermophile Determined At 1.55 A Resolution
Organism: Myceliophthora thermophila
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-01-02 Classification: HYDROLASE Ligands: EDO, GOL |
 |
Crystal Structure Of Glucuronoyl Esterase S213A Mutant From Sporotrichum Thermophile Determined At 1.9 A Resolution
Organism: Myceliophthora thermophila
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-01-02 Classification: HYDROLASE Ligands: EDO, GOL |

