Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2023-12-20 Classification: HYDROLASE Ligands: NAG, GOL, PEG, PG4, 1PE |
 |
Structure Of Prolyl Endoprotease From Aspergillus Niger Cbs 109712 In Space Group C222(1)
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-12-20 Classification: HYDROLASE Ligands: PG4 |
 |
Transcriptional Regulator Lmrr With Trp-67 And Trp-96 Replaced By The Unnatural Amino Acid 5-Fluorotrp
Organism: Lactococcus cremoris
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-11-23 Classification: DNA BINDING PROTEIN |
 |
Transcriptional Regulator Lmrr With Bound Daunomycin And With Trp-67 And Trp-96 Replaced By 5-Fluorotrp
Organism: Lactococcus cremoris
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-11-23 Classification: DNA BINDING PROTEIN Ligands: DM1 |
 |
Transcriptional Regulator Lmrr With Bound Daunomycin And With Trp-67 And Trp-96 Replaced By 5,6,7-Trifluorotrp
Organism: Lactococcus cremoris
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-11-23 Classification: DNA BINDING PROTEIN Ligands: DM1 |
 |
Transcriptional Regulator Lmrr With Bound Daunomycin And With Trp-67 And Trp-96 Replaced By The Unnatural Amino Acid 5,6-Difluorotrp
Organism: Lactococcus cremoris
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-11-23 Classification: DNA BINDING PROTEIN Ligands: DM1 |
 |
Transcriptional Regulator Lmrr With Trp-67 And Trp-96 Replaced By The Unnatural Amino Acid 5,6-Difluorotrp
Organism: Lactococcus cremoris
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2022-11-23 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of A Thermostable Glycoside Hydrolase Family 43 {Beta}-1,4-Xylosidase From Geobacillus Thermoleovorans It-08
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-04-25 Classification: HYDROLASE Ligands: CA, GOL |
 |
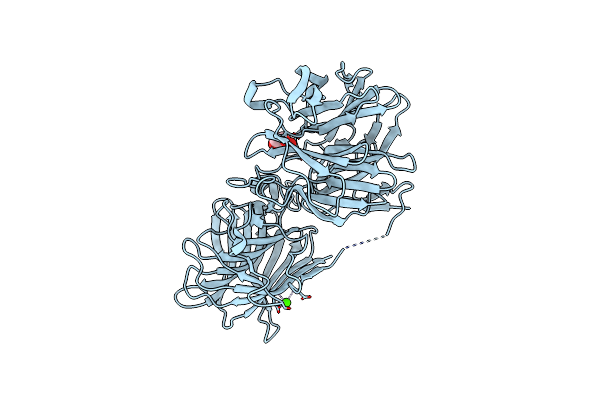
Crystal Structure Of A Thermostable Glycoside Hydrolase Family 43 {Beta}-1,4-Xylosidase From Geobacillus Thermoleovorans It-08 In Complex With L-Arabinose
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-04-25 Classification: HYDROLASE Ligands: CA, FUB |
 |
Crystal Structure Of A Thermostable Glycoside Hydrolase Family 43 {Beta}-1,4-Xylosidase From Geobacillus Thermoleovorans It-08 In Complex With D-Xylose
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-04-25 Classification: HYDROLASE Ligands: CA, XYS |
 |
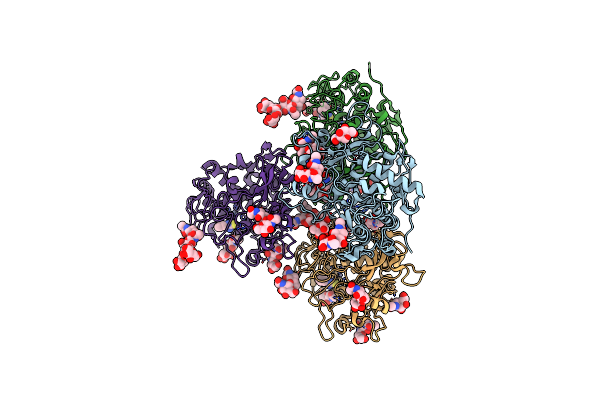
Crystal Structure Of A Thermostable Glycoside Hydrolase Family 43 {Beta}-1,4-Xylosidase From Geobacillus Thermoleovorans It-08 In Complex With L-Arabinose And D-Xylose
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-04-25 Classification: HYDROLASE Ligands: CA, FUB, XYS, XYP |
 |
Crystal Structure Of Human Tyrosinase Related Protein 1 Mutant (T391V-R374S-Y362F) In Complex With Phenylthiourea (Ptu)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-01-31 Classification: UNKNOWN FUNCTION Ligands: NAG, URS, ZN |
 |
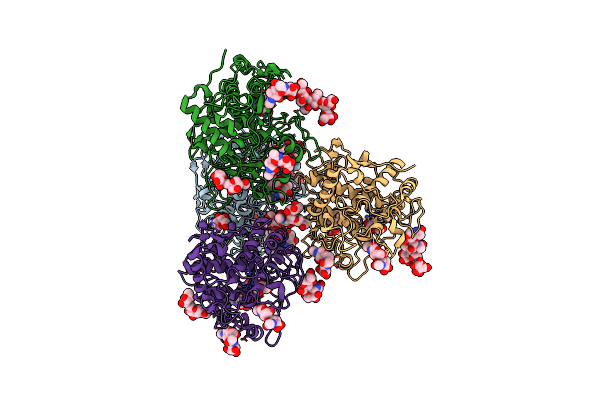
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-07-12 Classification: UNKNOWN FUNCTION Ligands: ZN, NAG |
 |
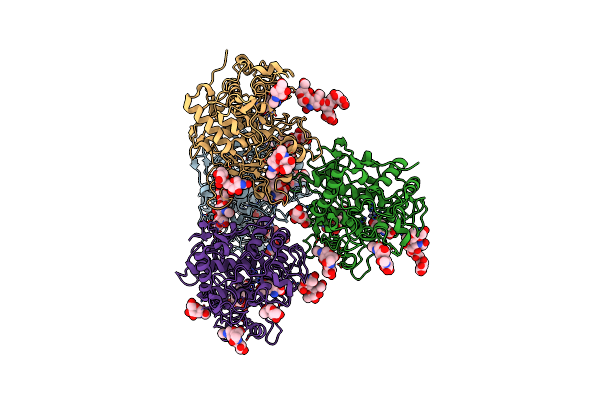
Crystal Structure Of Human Tyrosinase Related Protein 1 In Complex With Kojic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2017-07-12 Classification: UNKNOWN FUNCTION Ligands: ZN, KOJ, NAG |
 |
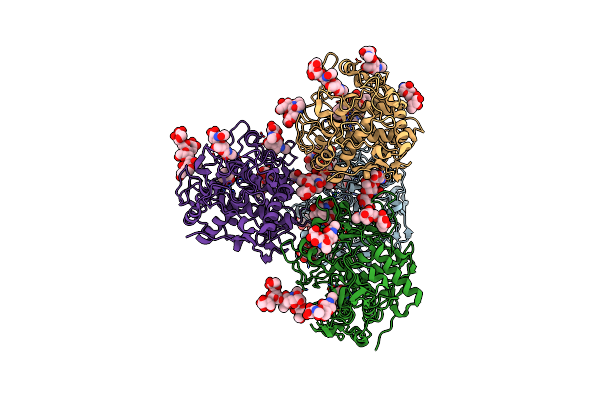
Crystal Structure Of Human Tyrosinase Related Protein 1 In Complex With Mimosine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-07-12 Classification: UNKNOWN FUNCTION Ligands: NAG, ZN, MMS |
 |
Crystal Structure Of Human Tyrosinase Related Protein 1 In Complex With Tropolone
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-07-12 Classification: UNKNOWN FUNCTION Ligands: 0TR, ZN |
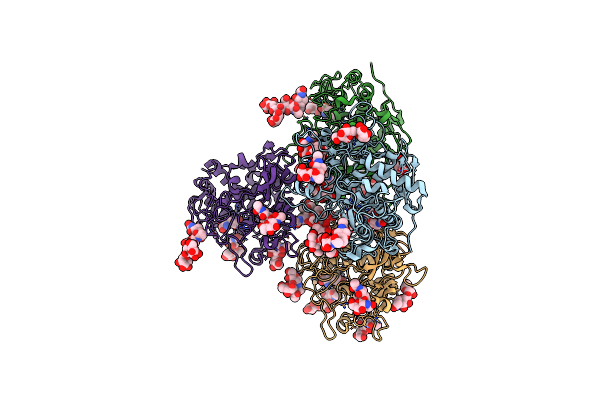 |
Crystal Structure Of Human Tyrosinase Related Protein 1 In Complex With Tyrosine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-07-12 Classification: UNKNOWN FUNCTION Ligands: NAG, TYR, ZN, MAN |
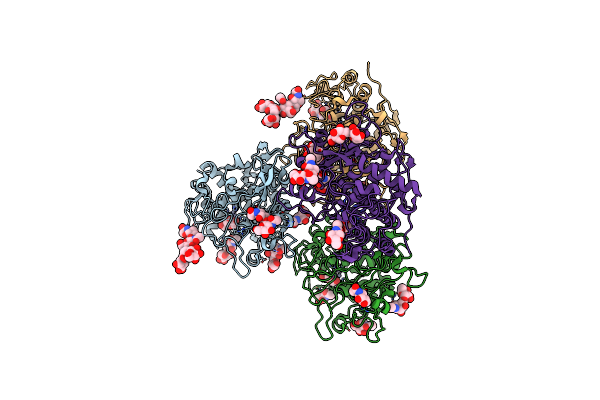 |
Crystal Structure Of Human Tyrosinase Related Protein 1 Mutant (T391V-R374S-Y362F) In Complex With Kojic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2017-07-12 Classification: UNKNOWN FUNCTION Ligands: NAG, ZN, KOJ |
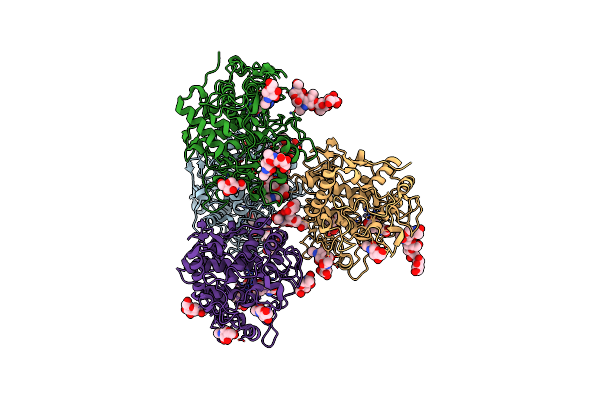 |
Crystal Structure Of Human Tyrosinase Related Protein 1 (T391V-R374S-Y362F) In Complex With Mimosine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-07-12 Classification: UNKNOWN FUNCTION Ligands: NAG, ZN, MMS |
 |
Crystal Structure Of Human Tyrosinase Related Protein 1 (T391V-R374S-Y362F) In Complex With Tropolone
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-07-12 Classification: UNKNOWN FUNCTION Ligands: NAG, ZN, 0TR |