Search Count: 13
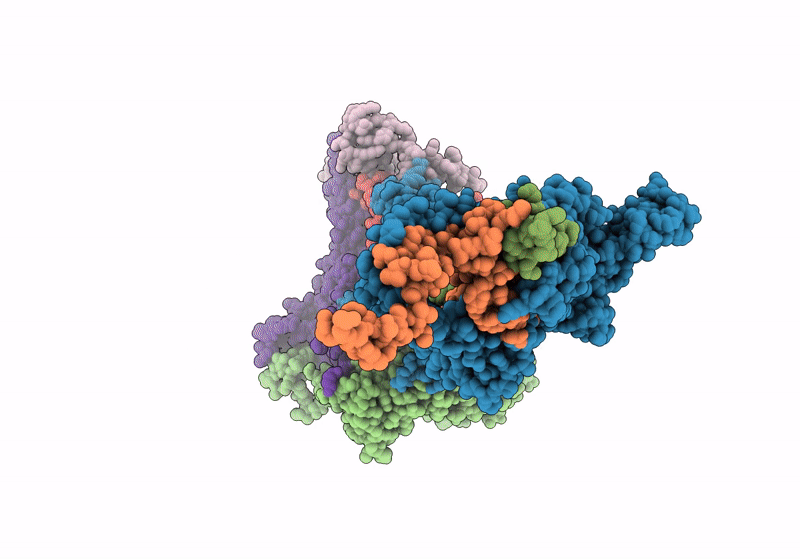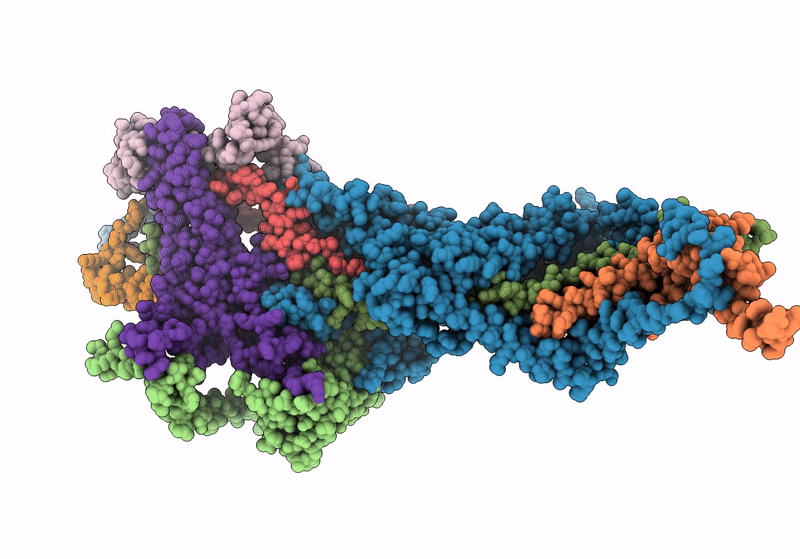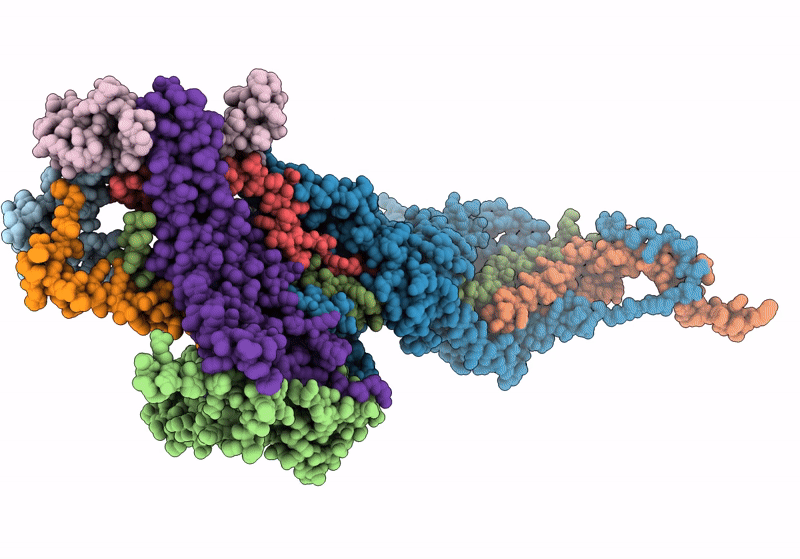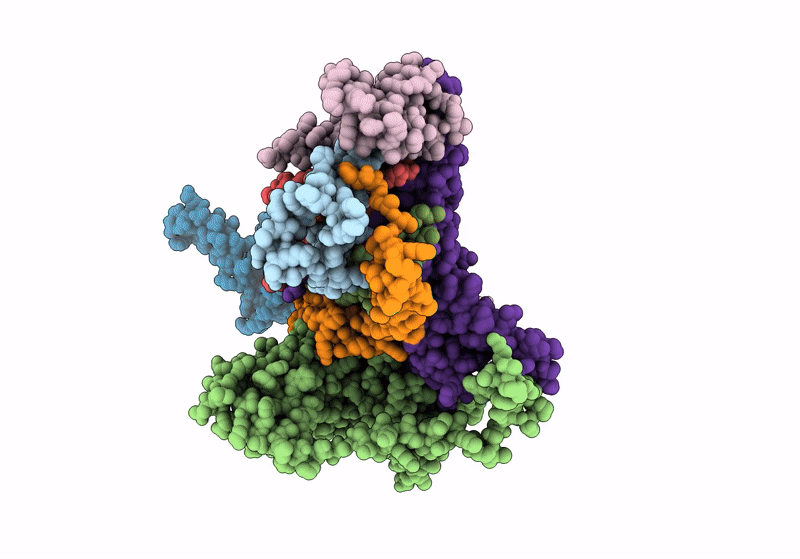 |
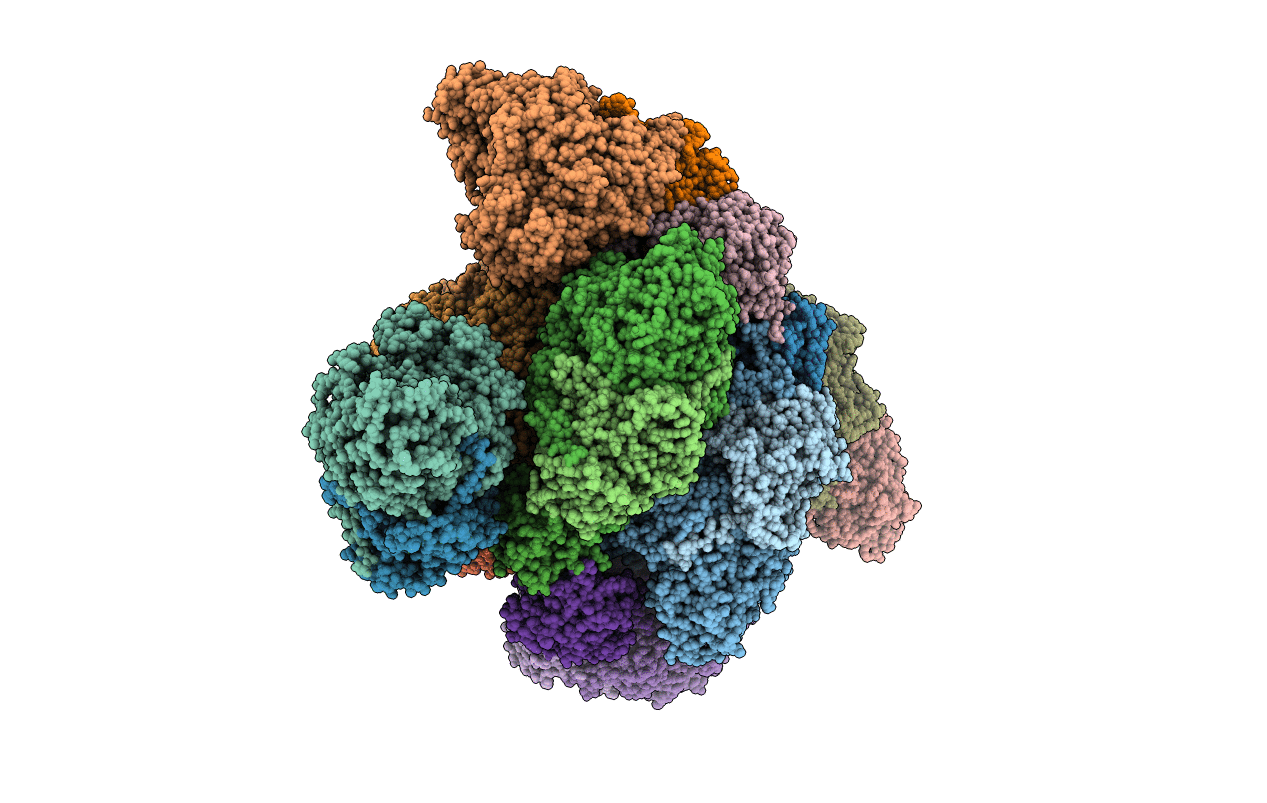
In Situ Structure Of The Peripheral Stalk Of The Mitochondrial Atpsynthase In Whole Polytomella Cells
Organism: Polytomella sp. pringsheim 198.80
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: MEMBRANE PROTEIN |
 |
Structure Of Nitrite Oxidoreductase (Nxr) From The Anammox Bacterium Kuenenia Stuttgartiensis.
Organism: Kuenenia stuttgartiensis
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2021-07-14 Classification: OXIDOREDUCTASE Ligands: SF4, F3S, MD1, MO, HEM, CA |
 |
Structure Of S-Mgm1 Decorating The Outer Surface Of Tubulated Lipid Membranes
Organism: Chaetomium thermophilum
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: CONTRACTILE PROTEIN |
 |
Structure Of S-Mgm1 Decorating The Outer Surface Of Tubulated Lipid Membranes In The Gtpgammas Bound State
Organism: Chaetomium thermophilum var. thermophilum dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: CONTRACTILE PROTEIN |
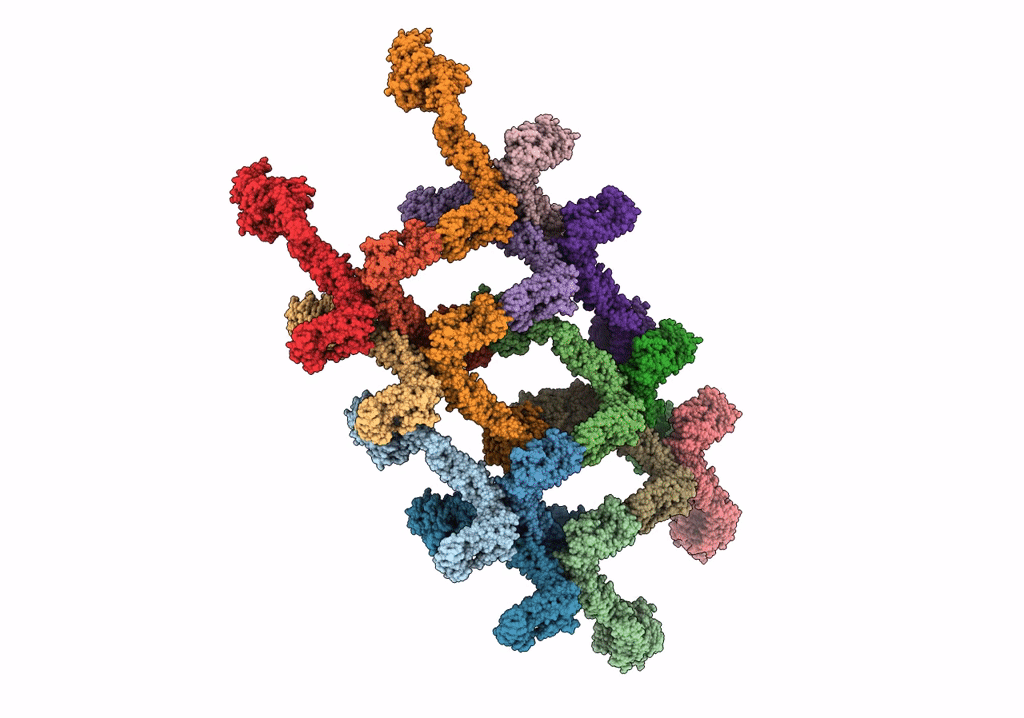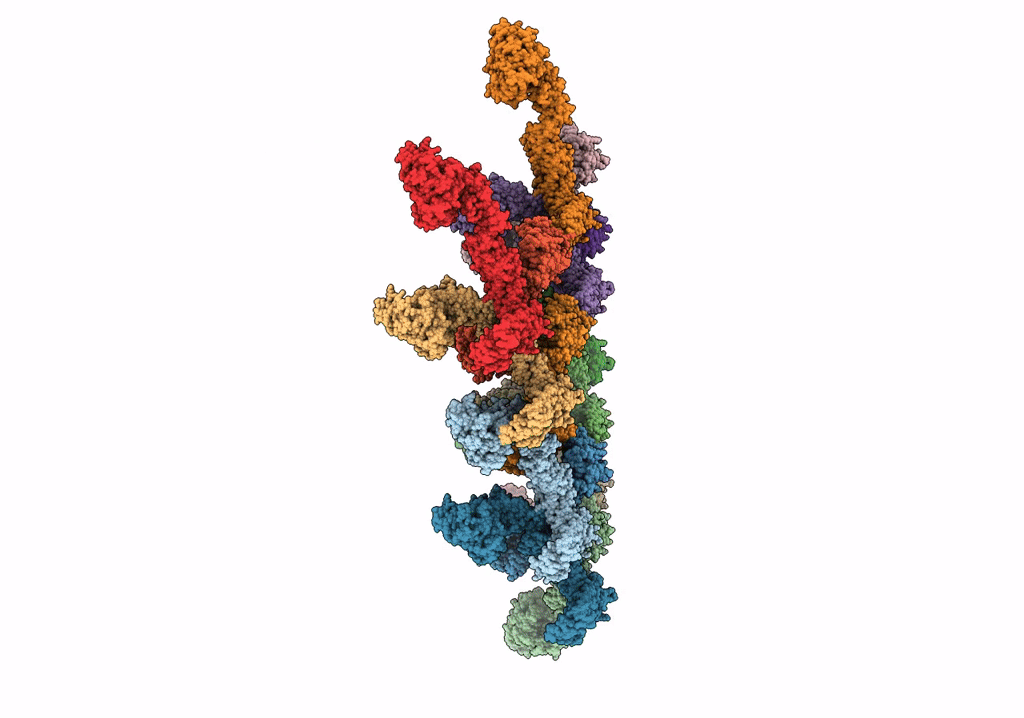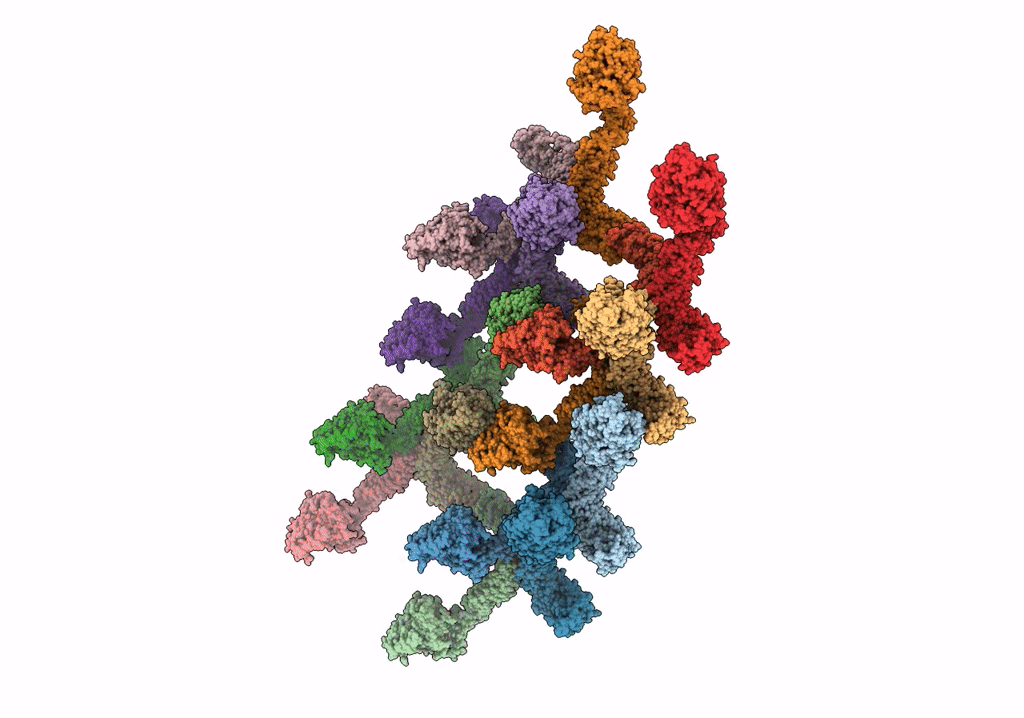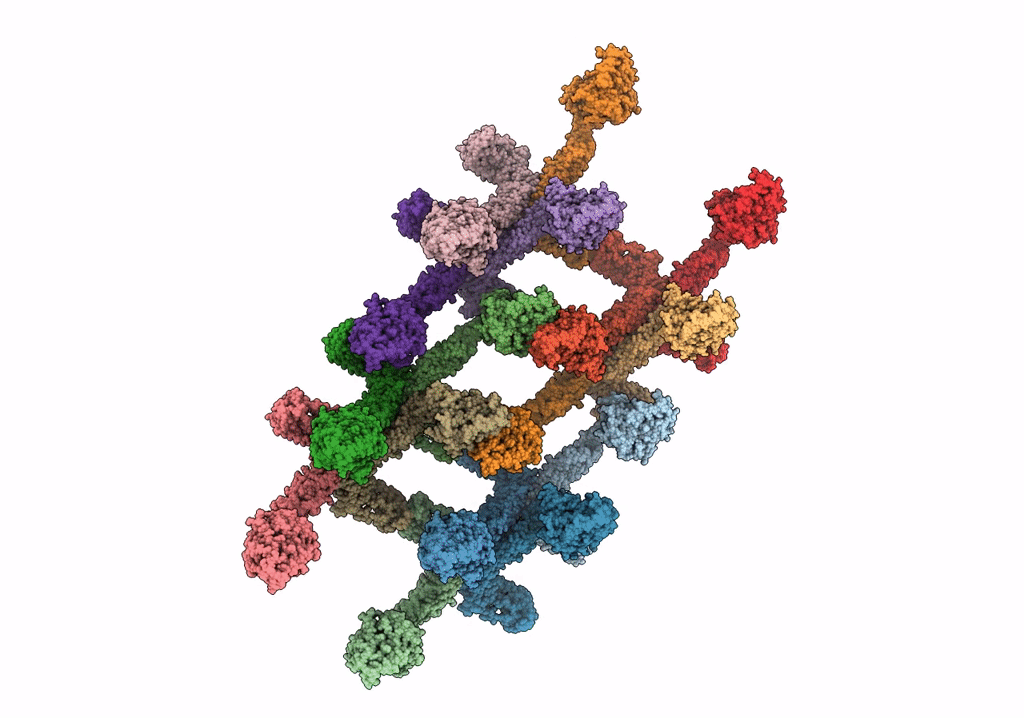 |
Structure Of S-Mgm1 Decorating The Inner Surface Of Tubulated Lipid Membranes
Organism: Chaetomium thermophilum var. thermophilum dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: CONTRACTILE PROTEIN |
 |
Structure Of S-Mgm1 Decorating The Inner Surface Of Tubulated Lipid Membranes In The Gtpgammas Bound State
Organism: Chaetomium thermophilum var. thermophilum dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: CONTRACTILE PROTEIN |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2019-07-03 Classification: MOTOR PROTEIN Ligands: EDO |
 |
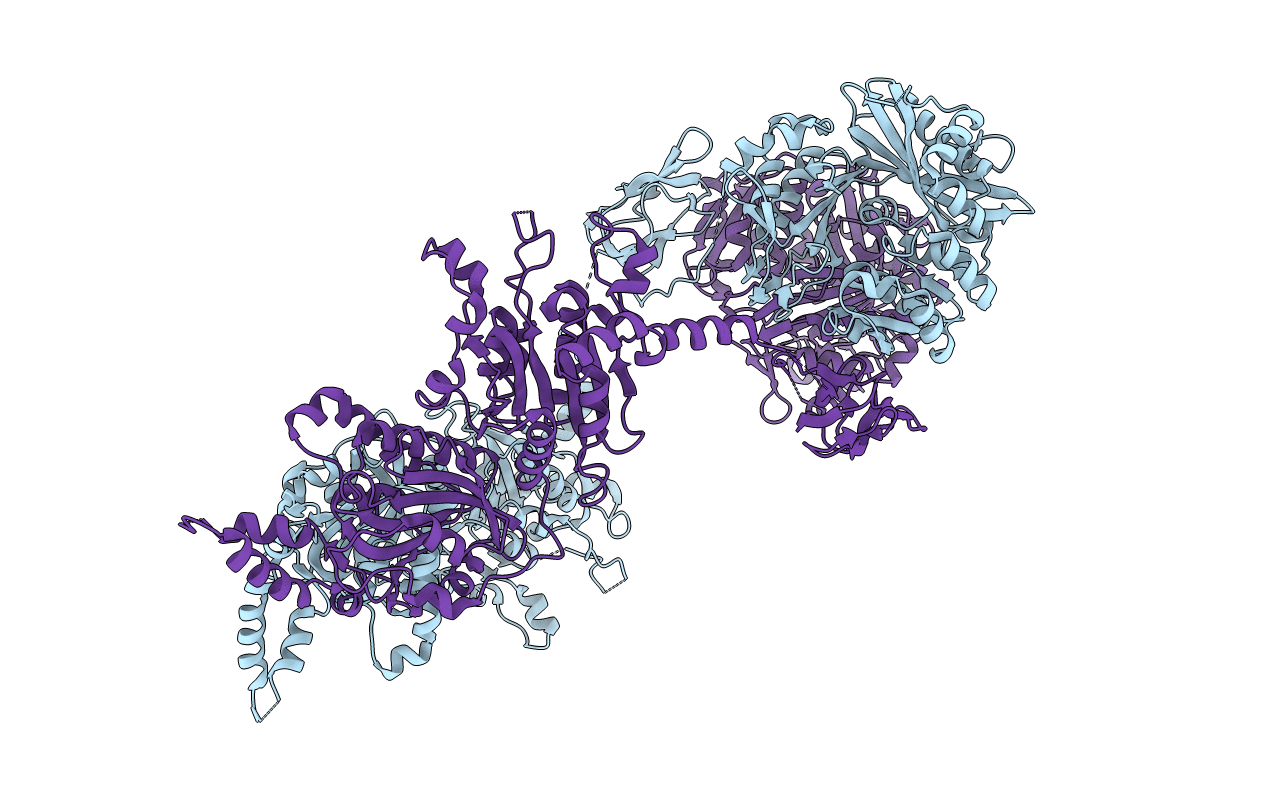
Crystal Structure Of Pseudomonas Malonate Decarboxylase Mdcd-Mdce Hetero-Dimer
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-08-16 Classification: TRANSFERASE |
 |
Crystal Structure Of A Pseudomonas Malonate Decarboxylase Hetero-Tetramer In Complex With Malonate
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1), Pseudomonas fluorescens (strain atcc baa-477 / nrrl b-23932 / pf-5), Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-08-16 Classification: TRANSFERASE Ligands: MLI |
 |
Crystal Structure Of A Pseudomonas Malonate Decarboxylase Hetero-Tetramer In Complex With Coenzyme A
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1), Pseudomonas fluorescens (strain atcc baa-477 / nrrl b-23932 / pf-5), Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-08-16 Classification: TRANSFERASE Ligands: CL, COA |
 |
Crystal Structure Of 14-3-3Zeta In Complex With A Cyclic Peptide Involving An Adamantyl And A Dicarboxy Side Chain
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2017-05-10 Classification: SIGNALING PROTEIN Ligands: BEZ |
 |
Crystal Structure Of Two-Subunit Pyruvate Carboxylase From Methylobacillus Flagellatus
Organism: Methylobacillus flagellatus, Methylobacillus flagellatus (strain kt / atcc 51484 / dsm 6875)
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2016-10-19 Classification: LIGASE Ligands: BTI, PYR, MN |
 |
Structure And Function Of A Single-Chain, Multi-Domain Long-Chain Acyl-Coa Carboxylase
Organism: Mycobacterium avium subsp. paratuberculosis
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2014-11-05 Classification: LIGASE |

