Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
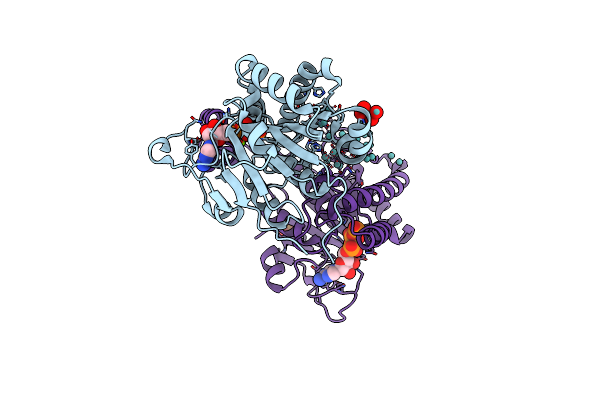 |
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2014-08-13 Classification: METAL BINDING PROTEIN Ligands: ATP, 8M0, MO, PO4, MG, M10 |
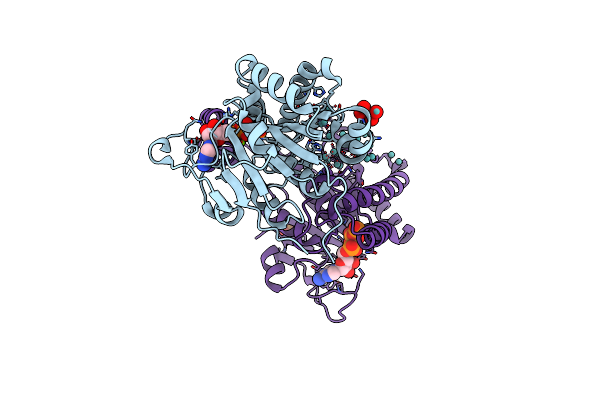 |
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-08-13 Classification: METAL BINDING PROTEIN Ligands: ATP, 8M0, MO, PO4, MG, M10 |
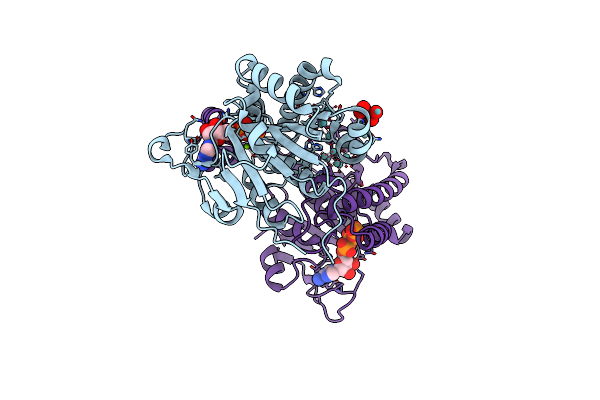 |
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-08-13 Classification: METAL BINDING PROTEIN Ligands: ATP, 8M0, MO, MG, M10 |
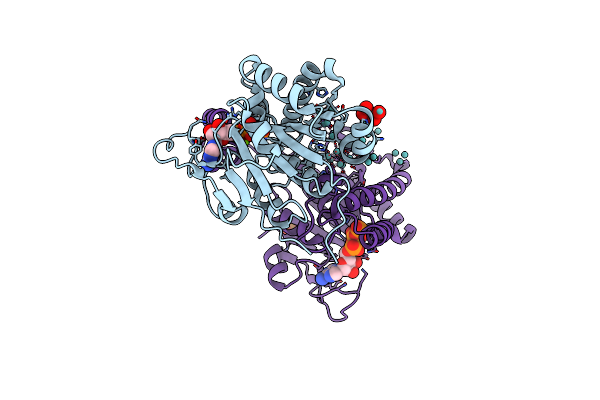 |
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-08-13 Classification: METAL BINDING PROTEIN Ligands: ATP, PO4, 8M0, MO, MG, M10 |
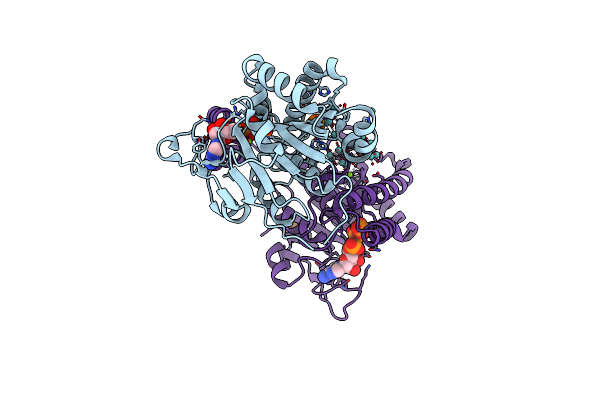 |
The Crystal Structure Of The Molybdenum Storage Protein (Mosto) From Azotobacter Vinelandii Loaded With Various Polyoxometalates
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-07-18 Classification: METAL BINDING PROTEIN Ligands: ATP, 8M0, PO4, MG, 6M0, MO |
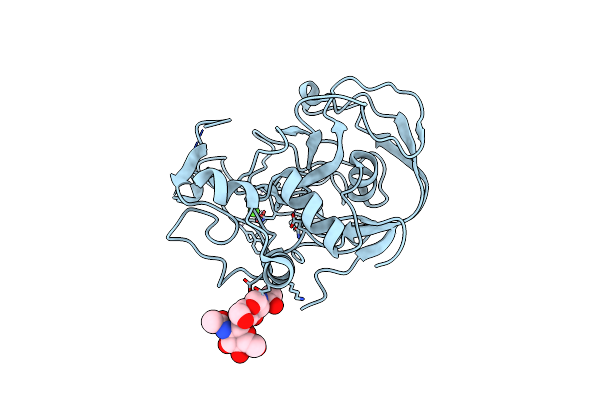 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2005-12-13 Classification: HYDROLASE ACTIVATOR, PROTEIN BINDING Ligands: CA |
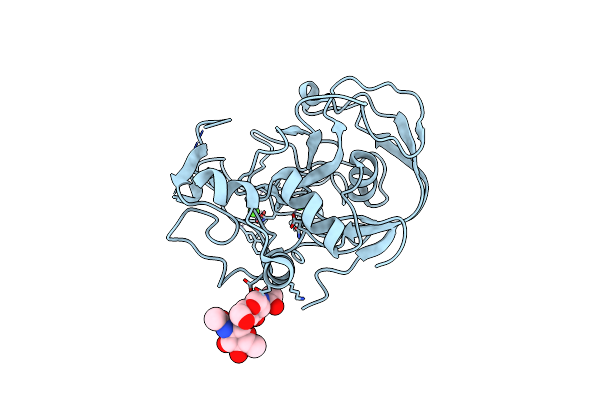 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2005-12-13 Classification: HYDROLASE ACTIVATOR, PROTEIN BINDING Ligands: CA |
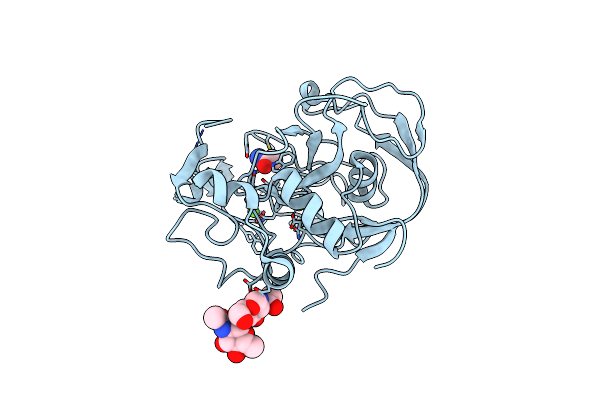 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2005-12-13 Classification: HYDROLASE ACTIVATOR,PROTEIN BINDING Ligands: CA, ACM |
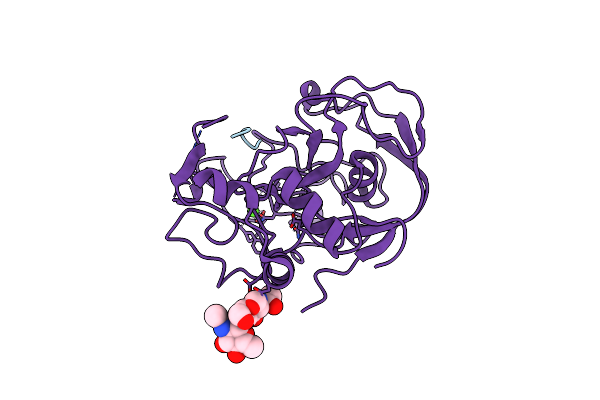 |
Formylglycine Generating Enzyme C336S Mutant Covalently Bound To Substrate Peptide Ctpsr
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2005-12-13 Classification: HYDROLASE ACTIVATOR,PROTEIN BINDING Ligands: CA, CL |
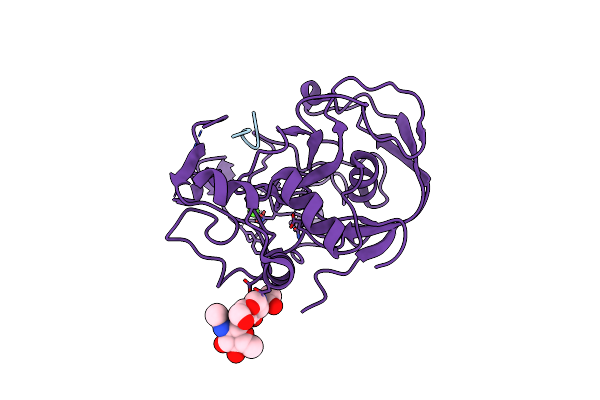 |
Formylglycine Generating Enzyme C336S Mutant Covalently Bound To Substrate Peptide Lctpsra
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2005-12-13 Classification: HYDROLASE ACTIVATOR,PROTEIN BINDING Ligands: CA, CL |
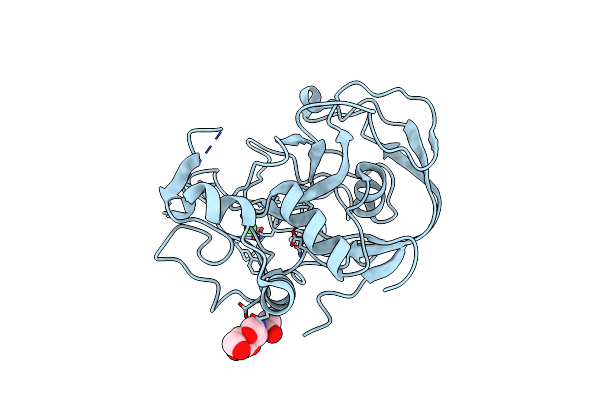 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2005-05-31 Classification: OXIDOREDUCTASE Ligands: NAG, CA |
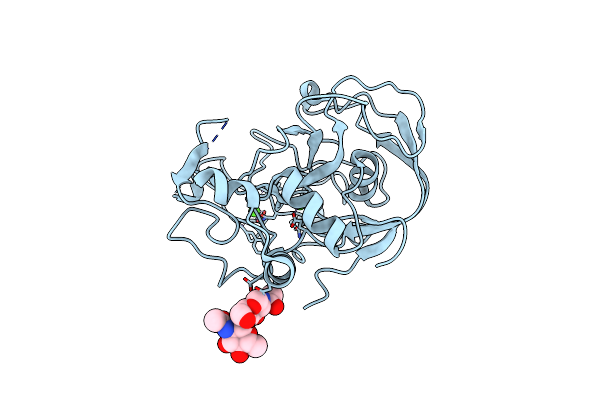 |
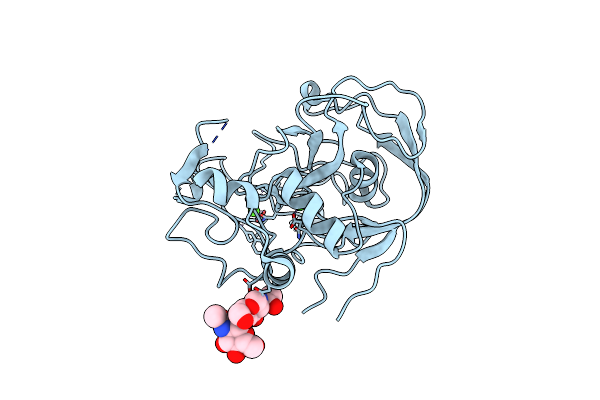
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-05-31 Classification: OXIDOREDUCTASE Ligands: CA |
 |
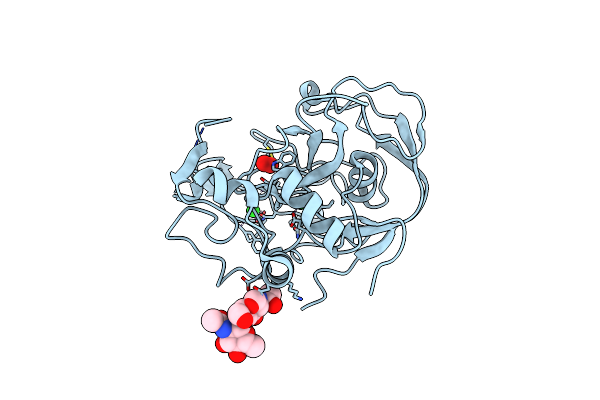
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2005-05-31 Classification: OXIDOREDUCTASE Ligands: CA |
 |
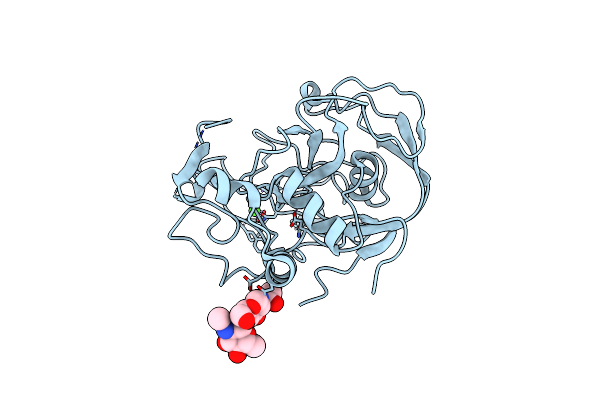
Human Formylglycine Generating Enzyme, Oxidised Cys Refined As Hydroperoxide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2005-05-31 Classification: OXIDOREDUCTASE Ligands: SR, PEO |
 |
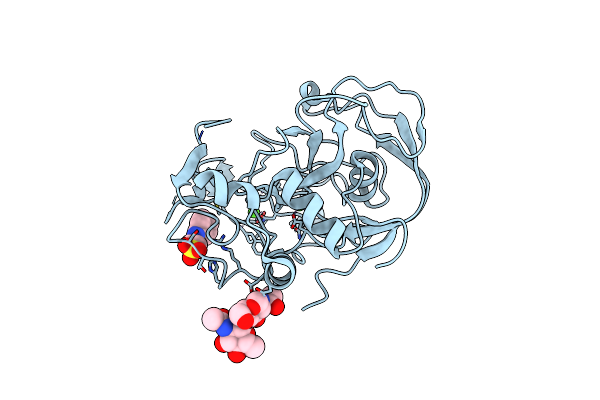
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2005-05-31 Classification: OXIDOREDUCTASE Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2005-05-31 Classification: OXIDOREDUCTASE Ligands: CA, CXS |
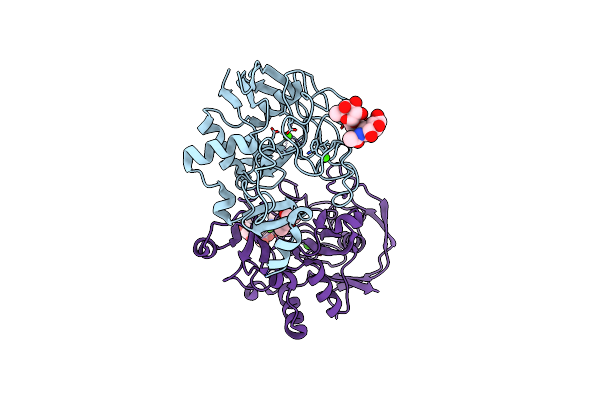 |
Crystal Structure Of The Paralogue Of The Human Formylglycine Generating Enzyme
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2005-02-08 Classification: SUGAR BINDING PROTEIN Ligands: CA, MPD |
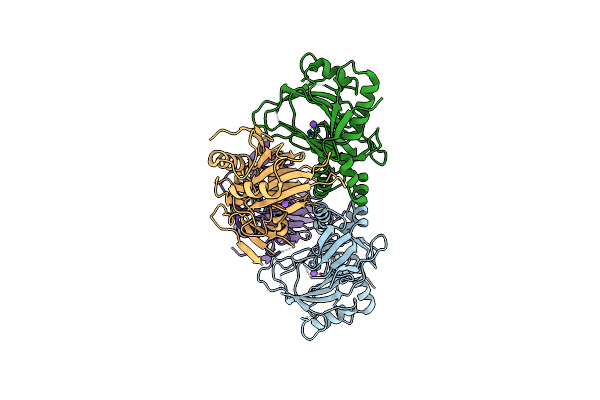 |
Crystal Structure Of The Alkylsulfatase Atsk, A Non-Heme Fe(Ii) Alphaketoglutarate Dependent Dioxygenase
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2004-03-30 Classification: OXIDOREDUCTASE |
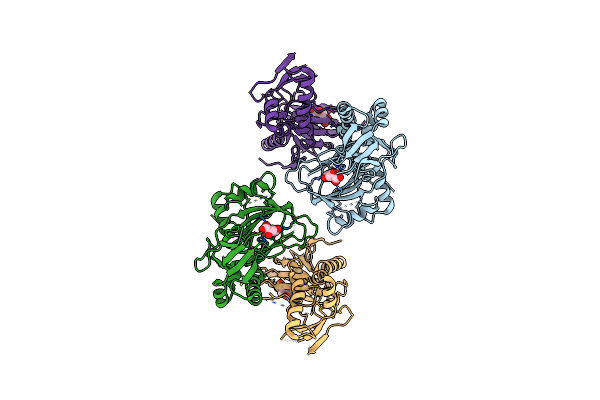 |
Crystal Structure Of The Alkylsulfatase Atsk, A Non-Heme Fe(Ii) Alphaketoglutarate Dependent Dioxygenase In Complex With Iron And Alphaketoglutarate
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2004-03-30 Classification: OXIDOREDUCTASE Ligands: FE2, AKG |
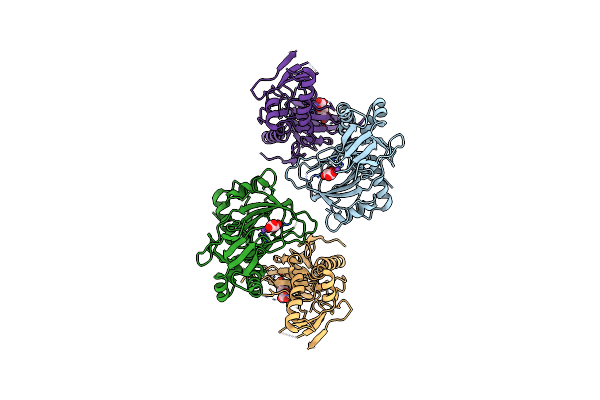 |
Crystal Structure Of The Alkylsulfatase Atsk, A Non-Heme Fe(Ii) Alphaketoglutarate Dependent Dioxygenase In Complex With Alphaketoglutarate
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-03-30 Classification: OXIDOREDUCTASE |