Search Count: 244
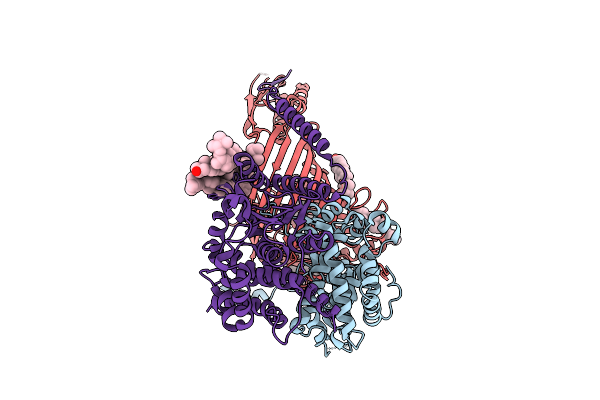 |
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
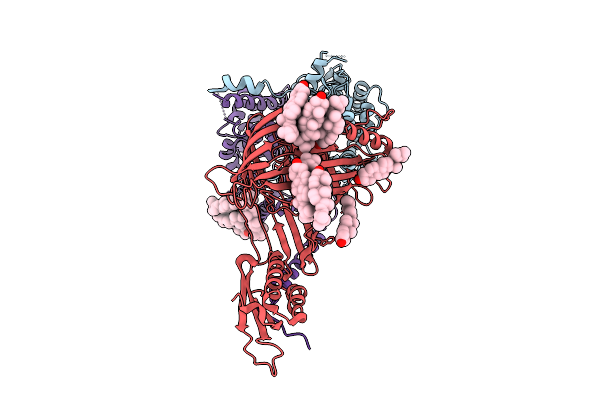 |
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
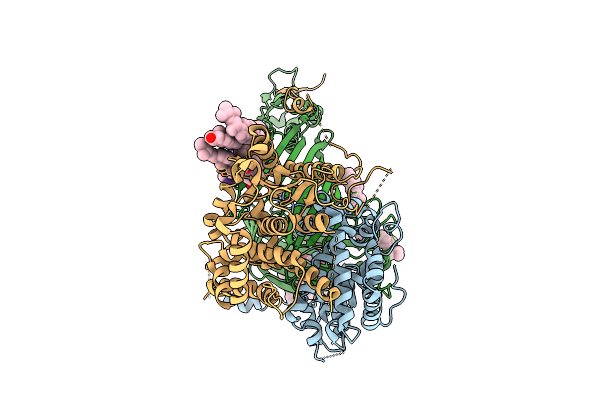 |
Organism: Thermothelomyces thermophilus, Photorhabdus khanii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN/ANTIBIOTIC Ligands: ERG |
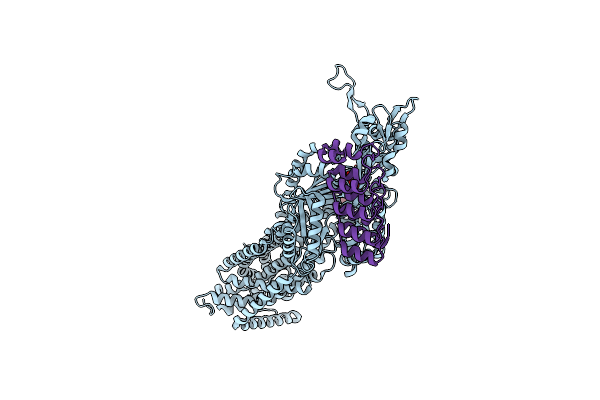 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN Ligands: MIY |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
Single Particle Cryo-Em Structure Of The Multidrug Efflux Pump Oqxb From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
Organism: Synthetic construct, Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN Ligands: MIY |
 |
Organism: Synthetic construct, Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:0.96 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: NO3, ACT |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: NO3, ACT |
 |
Neutron Structure Of Hydrogenated Hen Egg-White Lysozyme At Room Temperature
Organism: Gallus gallus
Method: NEUTRON DIFFRACTION Resolution:1.10 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: NO3, ACT, DOD |
 |
Neutron Structure Of Perdeuterated Hen Egg-White Lysozyme At Room Temperature
Organism: Gallus gallus
Method: NEUTRON DIFFRACTION Resolution:0.91 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: NO3, ACT, DOD |
 |
Cryo-Em Map Of The Focused Refinement Of The Subfamily Iii Haloalkane Dehalogenase From Haloferax Mediterranei Dimer Forming Hexameric Assembly.
Organism: Haloferax mediterranei
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: HYDROLASE |
 |
X-Ray Structure Of The Crystallization-Prone Form Of Subfamily Iii Haloalkane Dehalogenase Dhmea From Haloferax Mediterranei
Organism: Haloferax mediterranei
Method: X-RAY DIFFRACTION Resolution:3.31 Å Release Date: 2023-08-30 Classification: UNKNOWN FUNCTION Ligands: CL |
 |
Organism: Oryctolagus cuniculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-19 Classification: TRANSLATION Ligands: N, SPD, SPM, MG, UNX, GTP, AAC, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-04-12 Classification: STRUCTURAL PROTEIN Ligands: EDO |

