Search Count: 129
 |
Organism: Moesziomyces antarcticus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: CL, LYS |
 |
Crystal Structure Of A Double Mutant Of Virb8-Like Orfg Central And C-Terminal Domains Of Streptococcus Thermophilus Icest3 (Gram Positive Conjugative Type Iv Secretion System).
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-03-12 Classification: TRANSPORT PROTEIN |
 |
Glutathione Transferase Epsilon 1 From Drosophila Melanogaster In Complex With Glutathione
Organism: Drosophila melanogaster american nodavirus (anv) sw-2009a
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-09-04 Classification: TRANSFERASE Ligands: GOL, GSH, GS8 |
 |
Crystal Structure Of Rat Glutathione Transferase Omega 1 Bound To Glutathione
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-07-31 Classification: TRANSFERASE Ligands: GSH |
 |
Crystal Structure Of Apis Mellifera Glutathione Transferase Delta 1 In A Covalent Dimeric State
Organism: Apis mellifera
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-10-18 Classification: TRANSFERASE |
 |
Crystal Structure Of Apis Mellifera Glutathione Transferase Delta 1, Mutant M126L
Organism: Apis mellifera
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of Apis Mellifera Glutathione Transferase Delta 1, Mutant C127S
Organism: Apis mellifera
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of An Hexavariant Of The B1 Domain Of Human Neuropilin-1 In Complex With The Kdkppr Peptide
Organism: Homo sapiens, Unidentified
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-08-02 Classification: PROTEIN BINDING Ligands: ACT |
 |
Crystal Structure Of Glutathione Transferase Chi 1 From Synechocystis Sp. Pcc 6803 In Complex With Glutathione
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-31 Classification: TRANSFERASE Ligands: GSH |
 |
S10T Variant Of Glutathione Transferase Chi 1 From Synechocystis Sp. Pcc 6803 In Complex With Glutathione
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-31 Classification: TRANSFERASE Ligands: GSH |
 |
R11A Variant Of Glutathione Transferase Chi 1 From Synechocystis Sp. Pcc 6803 In Complex With Glutathione
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-31 Classification: TRANSFERASE Ligands: GSH |
 |
Crystal Structure Of Trametes Versicolor Glutathione Transferase Omega 3S In Complex With Dinitrosyl Glutathionyl Iron Complex (Dngic)
Organism: Trametes versicolor
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-03-01 Classification: TRANSFERASE Ligands: PG4, GOL, FE, NO, GSH, CA, PEG |
 |
Crystal Structure Of Trametes Versicolor Glutathione Transferase Omega 3S In Complex With Sodium Nitroprusside
Organism: Trametes versicolor
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-03-01 Classification: TRANSFERASE Ligands: NT3, ACT, GOL, PEG, CA |
 |
Crystal Structure Of Trametes Versicolor Glutathione Transferase Omega 3S In Complex With Hydroxy-Tetranitro-Nitrosyl-Ruthenate
Organism: Trametes versicolor
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-03-01 Classification: TRANSFERASE Ligands: ODU, ACT, GOL, RU, CA |
 |
Crystal Structure Of Trametes Versicolor Glutathione Transferase Omega 3S In Complex With Glutathione And Pentachloro-Nitrosyl-Osmate
Organism: Trametes versicolor
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-03-01 Classification: TRANSFERASE Ligands: OS, GOL, GSH, CA |
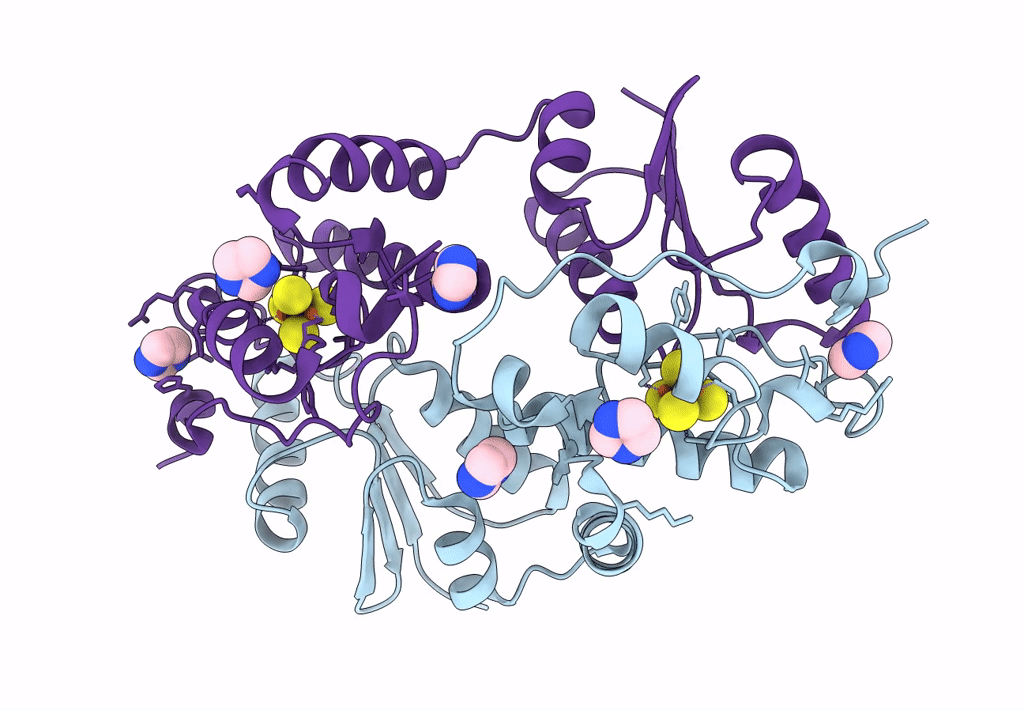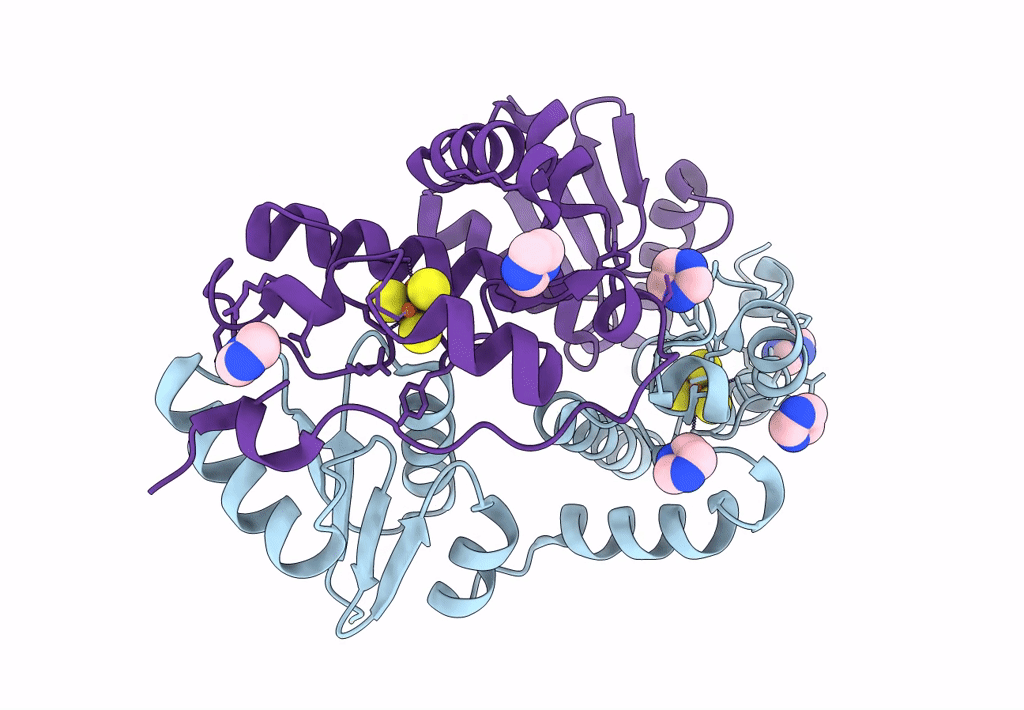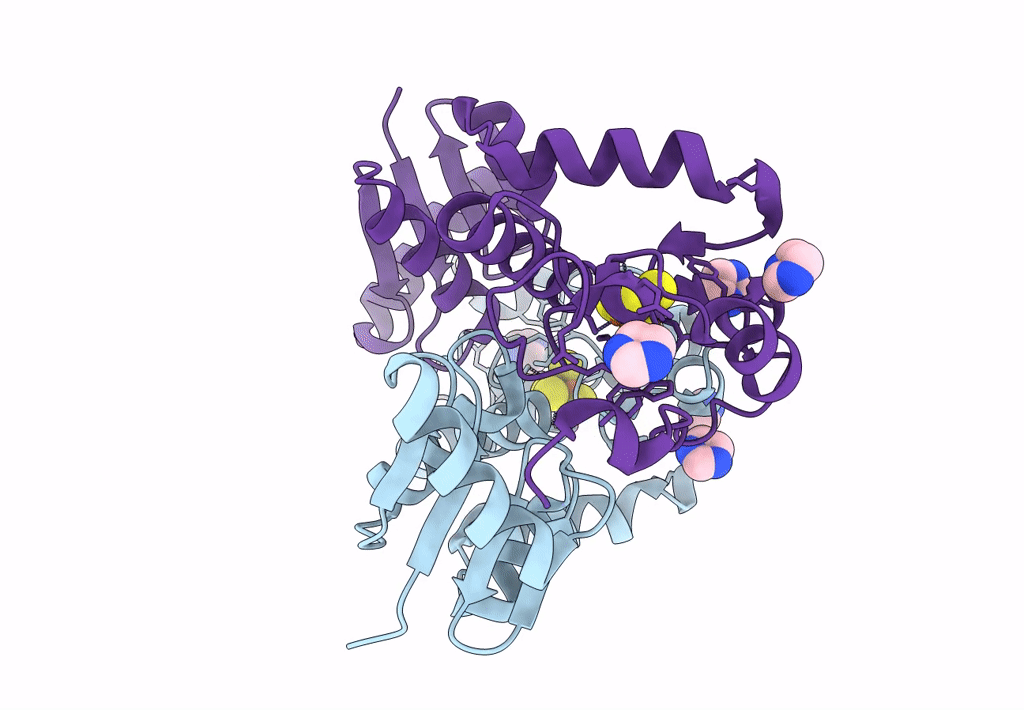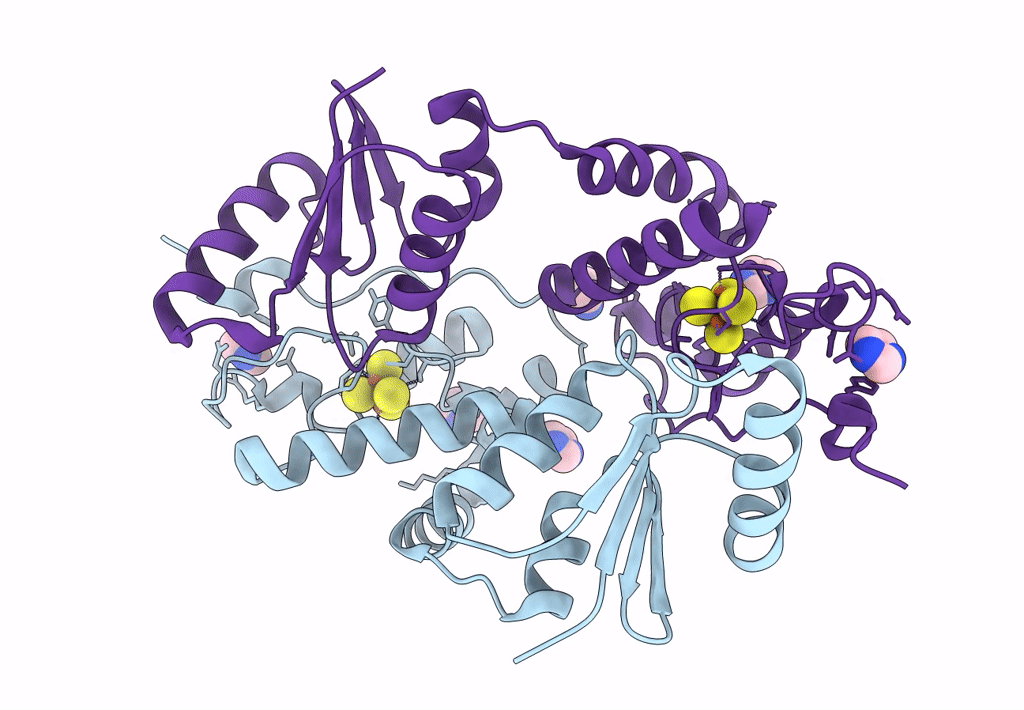 |
Crystal Structure Of The Glutaredoxin/Ferredoxin Disulfide Reductase Fusion Protein From Desulfotalea Psychrophila Lsv54
Organism: Desulfotalea psychrophila (strain lsv54 / dsm 12343)
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-10-05 Classification: UNKNOWN FUNCTION Ligands: SF4, IMD |
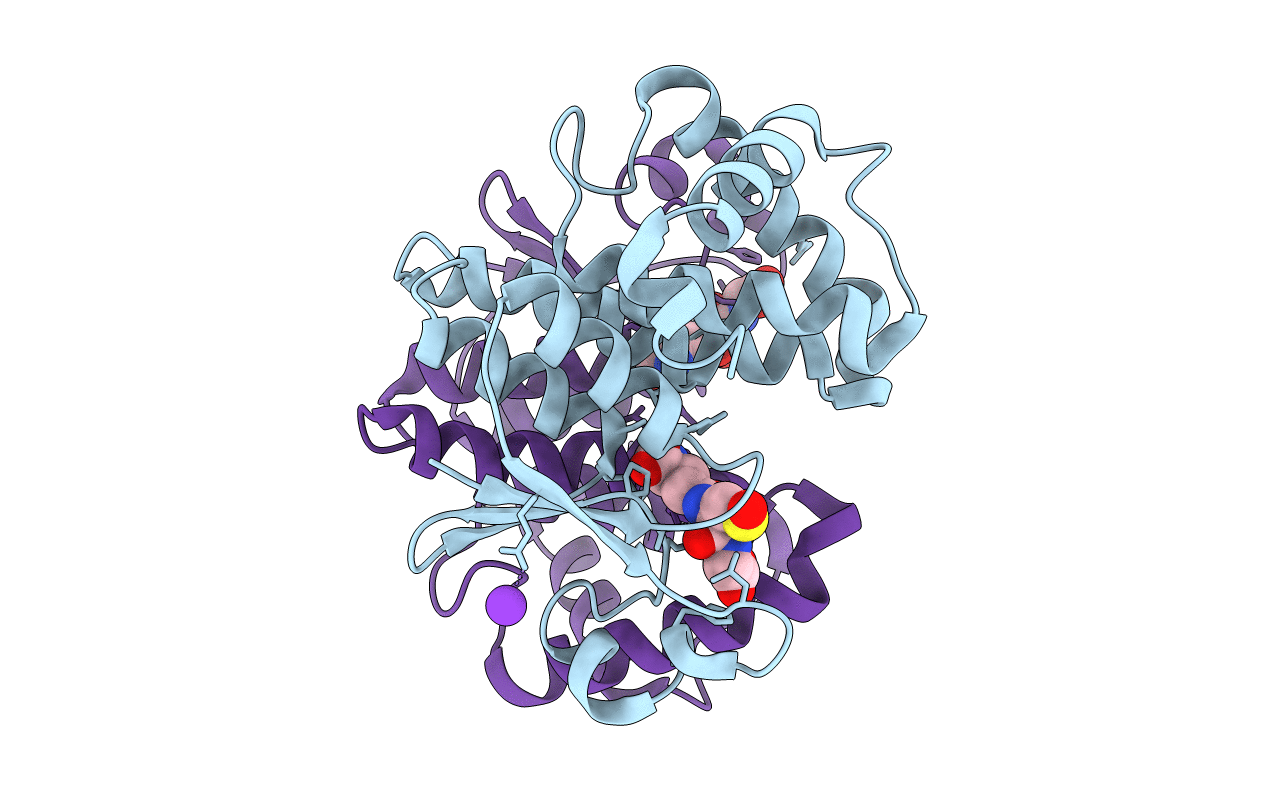 |
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2022-09-07 Classification: TRANSFERASE Ligands: GS8, K |
 |
Crystal Structure Of Virb8-Like Orfg Central And C-Terminal Domains Of Streptococcus Thermophilus Icest3 (Gram Positive Conjugative Type Iv Secretion System).
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-09-07 Classification: TRANSPORT PROTEIN Ligands: SO4, GOL |
 |
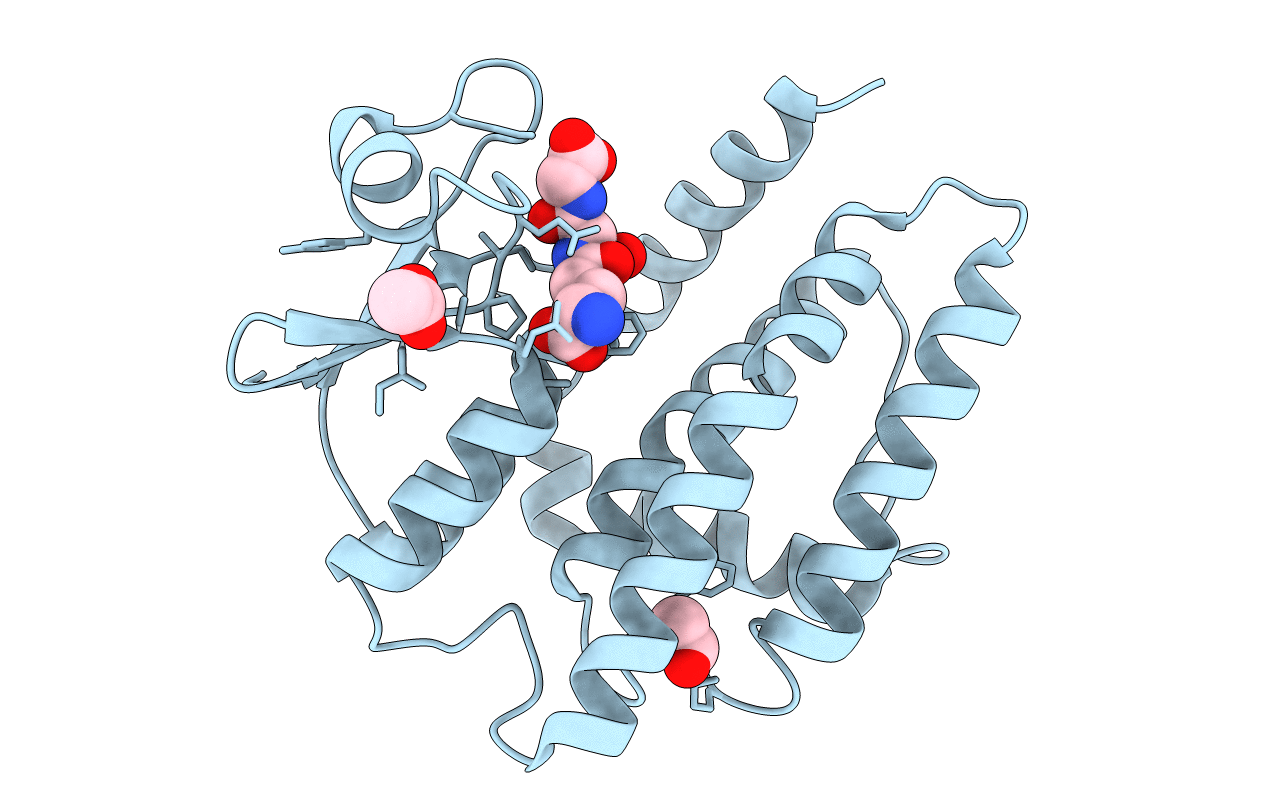
Organism: Populus trichocarpa
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-07-20 Classification: TRANSFERASE Ligands: ACT |
 |
Crystal Structure Of Poplar Glutathione Transferase U19 In Complex With Glutathione
Organism: Populus trichocarpa
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2022-07-20 Classification: TRANSFERASE Ligands: GS8, ACT |

