Search Count: 26
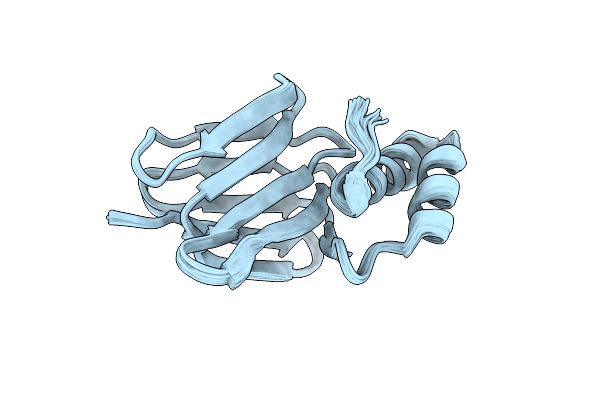 |
Solution Structure Of Subunit Epsilon Of The Mycobacterium Abscessus F-Atp Synthase
Organism: Mycobacteroides abscessus
Method: SOLUTION NMR Release Date: 2023-03-08 Classification: ELECTRON TRANSPORT |
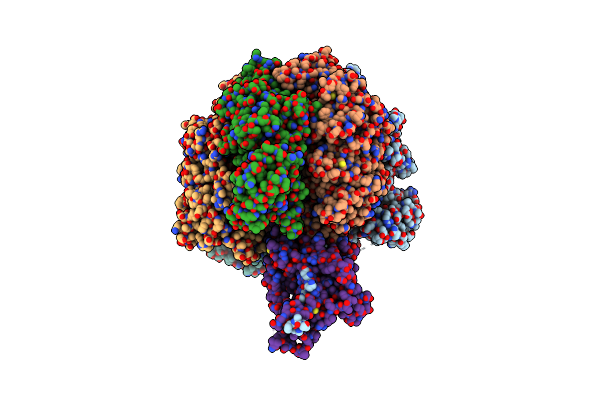 |
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: HYDROLASE Ligands: ATP, MG, ADP |
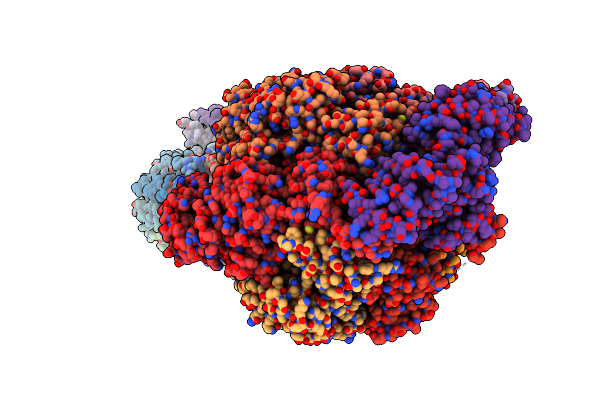 |
Cryo-Em Structure Of F-Atp Synthase From Mycolicibacterium Smegmatis (Rotational State 1)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: HYDROLASE Ligands: ATP, MG, ADP |
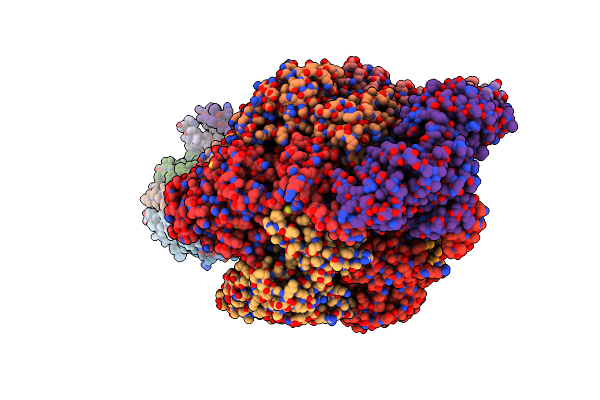 |
Cryo-Em Structure Of F-Atp Synthase From Mycolicibacterium Smegmatis (Rotational State 2)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: HYDROLASE Ligands: MG, ATP, ADP |
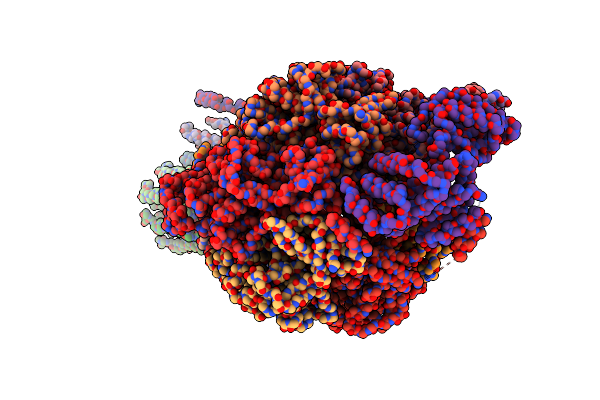 |
Cryo-Em Structure Of F-Atp Synthase From Mycolicibacterium Smegmatis (Rotational State 3) (Backbone)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: HYDROLASE |
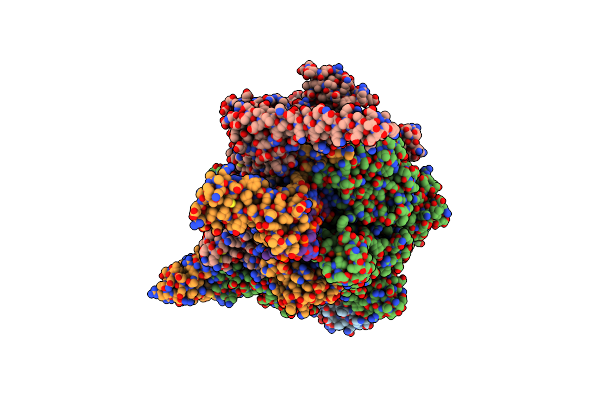 |
Mycobacterium Tuberculosis Rna Polymerase Sigma A Holoenzyme Open Promoter Complex Containing Umn-7
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:3.87 Å Release Date: 2022-10-19 Classification: ANTIBIOTIC Ligands: KYO, ZN, MG |
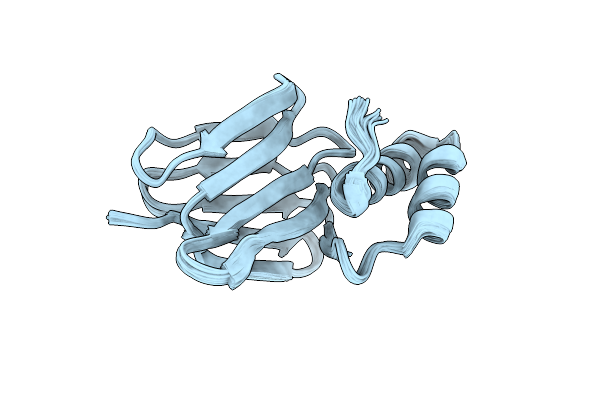 |
Solution Structure Of Subunit Epsilon Of The Mycobacterium Abscessus F-Atp Synthase
Organism: Mycobacteroides abscessus
Method: SOLUTION NMR Release Date: 2022-10-05 Classification: BIOSYNTHETIC PROTEIN |
 |
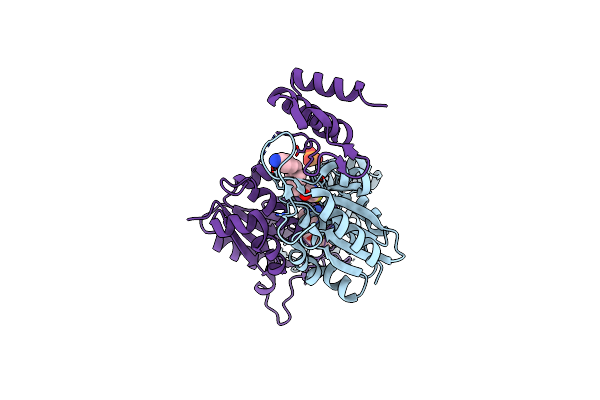
Structure Of F420 Binding Protein Rv1558 From Mycobacterium Tuberculosis With F420 Bound
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2021-03-24 Classification: OXIDOREDUCTASE Ligands: 6J4, SO4, F42 |
 |
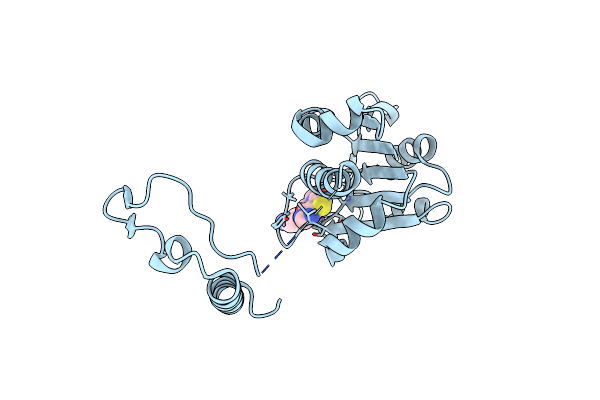
Crystal Structure Of Trmd From Pseudomonas Aeruginosa In Complex With Active-Site Inhibitor
Organism: Pseudomonas aeruginosa ucbpp-pa14
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2019-09-04 Classification: TRANSFERASE Ligands: BWR, PO4, SAM |
 |
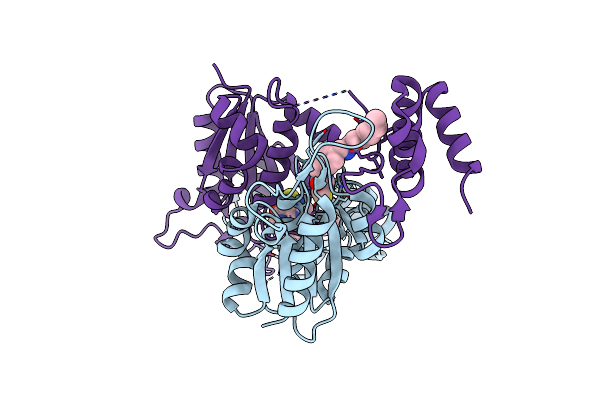
Crystal Structure Of Trmd From Mycobacterium Tuberculosis In Complex With Active-Site Inhibitor
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-09-04 Classification: TRANSFERASE/INHIBITOR Ligands: BWR |
 |
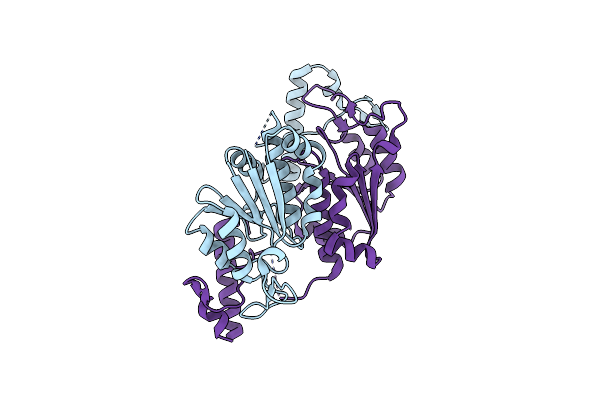
Crystal Structure Of Trmd From Pseudomonas Aeruginosa In Complex With Active-Site Inhibitor
Organism: Pseudomonas aeruginosa ucbpp-pa14
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2019-03-27 Classification: TRANSFERASE Ligands: 9D0 |
 |
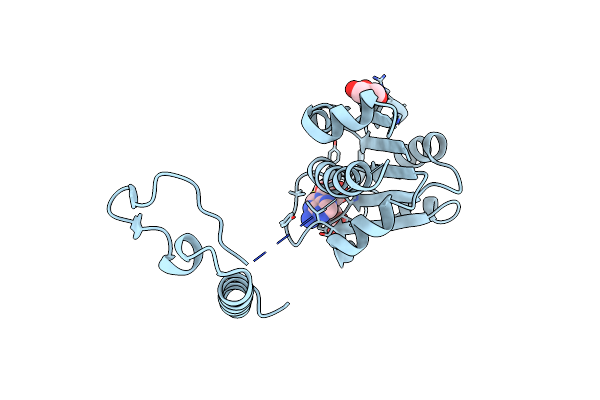
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-03-06 Classification: TRANSFERASE |
 |
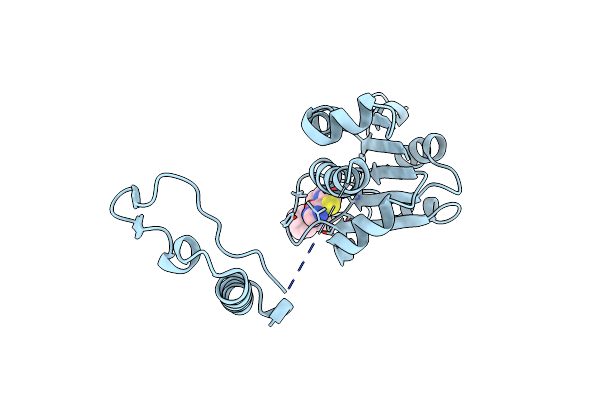
Crystal Structure Of Trmd From Mycobacterium Tuberculosis In Complex With S-Adenosyl Homocysteine (Sah)
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-03-06 Classification: TRANSFERASE Ligands: SAH, GOL |
 |
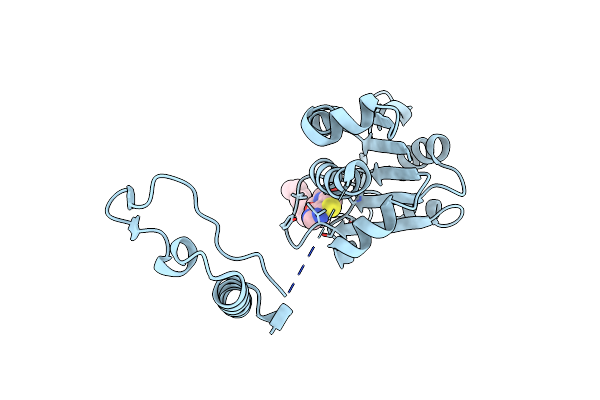
Crystal Structure Of Trmd From Mycobacterium Tuberculosis In Complex With Active-Site Inhibitor
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-03-06 Classification: TRANSFERASE Ligands: 9D6 |
 |
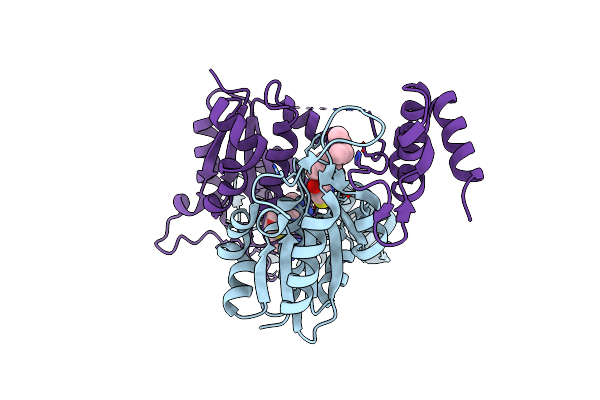
Crystal Structure Of Trmd From Mycobacterium Tuberculosis In Complex With Active-Site Inhibitor
Organism: Mycobacterium tuberculosis cdc 1551
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-03-06 Classification: TRANSFERASE Ligands: 9D0 |
 |
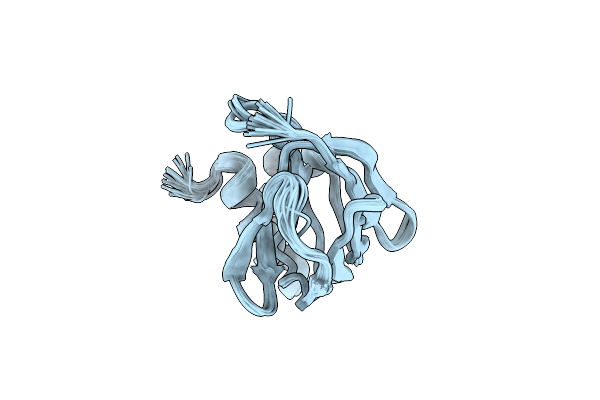
Crystal Structure Of Trmd From Pseudomonas Aeruginosa In Complex With Active-Site Inhibitor
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2019-03-06 Classification: TRANSFERASE Ligands: 9D3, SAM |
 |
Nmr Solution Structure Of Subunit Epsilon Of The Mycobacterium Tuberculosis F-Atp Synthase
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: SOLUTION NMR Release Date: 2018-10-10 Classification: PROTON TRANSPORT |
 |
Nmr Solution Structure Of Peptide Epsilon(103-120) From Mycobacterium Tuberculosis F-Atpsynthase
Organism: Mycobacterium tuberculosis
Method: SOLUTION NMR Release Date: 2012-11-07 Classification: HYDROLASE INHIBITOR |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: MES |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: SO4 |

