Search Count: 31
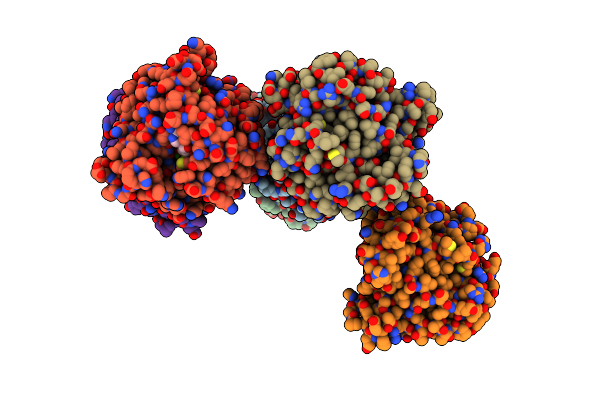 |
Crystal Structure Of Norbelladine O-Methyltransferase Variant In Complex With Sah
Organism: Narcissus aff. pseudonarcissus mk-2014
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-03-20 Classification: TRANSFERASE Ligands: SAH, CA |
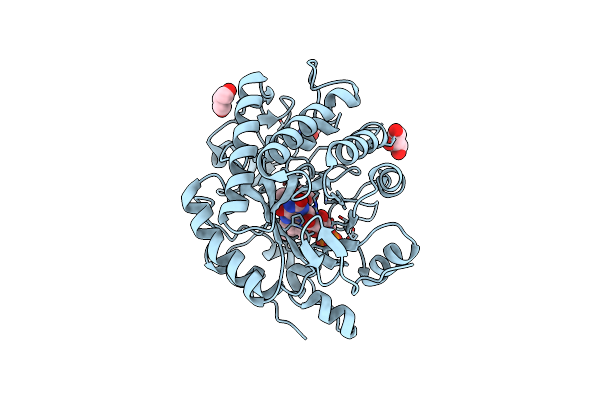 |
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2024-03-13 Classification: OXIDOREDUCTASE Ligands: ACT, GOL, FMN |
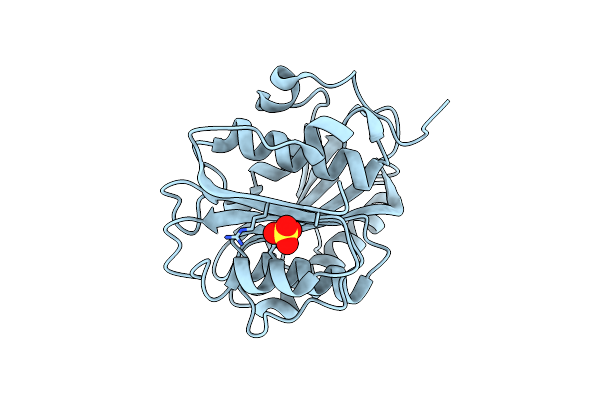 |
Organism: Ideonella sakaiensis
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2022-04-06 Classification: HYDROLASE Ligands: SO4 |
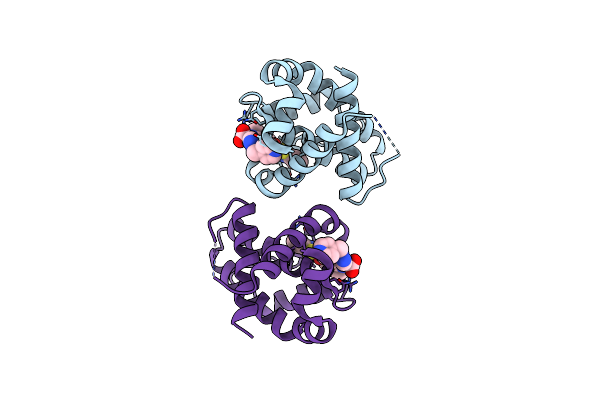 |
Crystal Structure Of Bcl-Xl In Complex With Tetrahydroisoquinoline-Pyridine Based Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-10-21 Classification: APOPTOSIS/APOPTOSIS INHIBITOR Ligands: RQ7 |
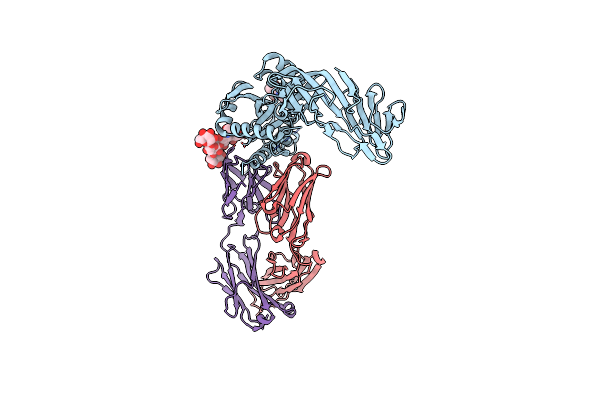 |
Crystal Structure Of A Human Metapneumovirus Monomeric Fusion Protein Complexed With 458 Fab
Organism: Human metapneumovirus, Human immunodeficiency virus 1, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-06-03 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Staphylococcus aureus (strain nctc 8325)
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2019-06-26 Classification: HYDROLASE Ligands: JT7 |
 |
3D Nmr Solution Structure Of Ligand Peptide (Ac)Evnppvp Of Pro-Pro Endopeptidase-1
Organism: Clostridioides difficile
Method: SOLUTION NMR Release Date: 2019-06-19 Classification: HYDROLASE |
 |
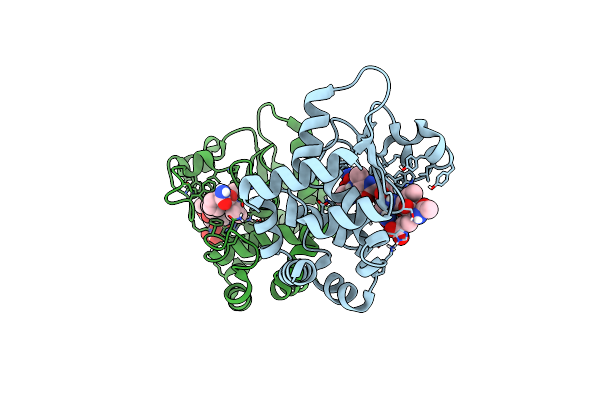
Crystal Structure Of Apo Ppep-1(E143A/Y178F) In Complex With Substrate Peptide Ac-Evnapvp-Conh2
Organism: Peptoclostridium difficile, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN |
 |
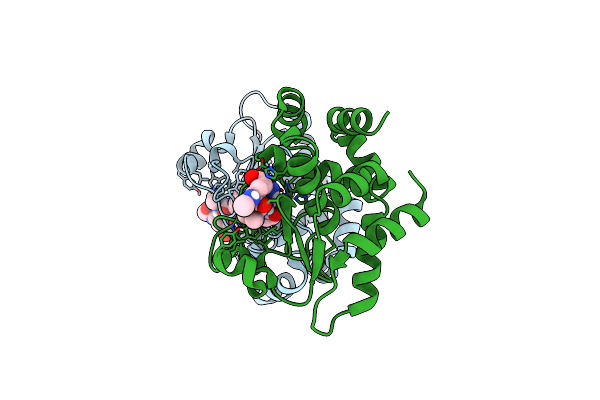
Crystal Structure Of Ppep-1(E143A/Y178F) In Complex With Substrate Peptide Ac-Evnpavp-Conh2
Organism: Peptoclostridium difficile, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Holo Ppep-1(E143A/Y178F) In Complex With Product Peptide Ac-Evnp-Co2 (Substrate Peptide: Ac-Evnpavp-Conh2)
Organism: Peptoclostridium difficile, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Holo Ppep-1(E143A/Y178F) In Complex With Product Peptide Ac-Evnp-Co2 (Substrate Peptide: Ac-Evnppvp-Conh2)
Organism: Peptoclostridium difficile, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN, NI |
 |
Crystal Structure Of Holo Ppep-1(E143A/Y178F) In Complex With Substrate Peptide Ac-Evnapvp-Conh2
Organism: Peptoclostridium difficile , Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Apo Ppep-1(E143A/Y178F) In Complex With Fibrinogen-Derived Substrate Peptide Ac-Slrpapp-Conh2
Organism: Peptoclostridium difficile, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: PO4 |
 |
Organism: Peptoclostridium difficile
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: TRS, ZN |
 |
Organism: Peptoclostridium difficile
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN, TRS |
 |
Organism: Peptoclostridium difficile
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: TRS, ZN, NI |
 |
Organism: Peptoclostridium difficile
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN, TRS |
 |
Organism: Peptoclostridium difficile
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN, TRS |
 |
Crystal Structure Of Ppep-1(E143A/Y178F/E184A) In Complex With Substrate Peptide Ac-Evnppvp-Conh2
Organism: Peptoclostridium difficile, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Ppep-1(E143A/Y178F/E184A) In Complex With Substrate Peptide Ac-Evnapvp-Conh2
Organism: Peptoclostridium difficile, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN, PO4, NI |

