Search Count: 117
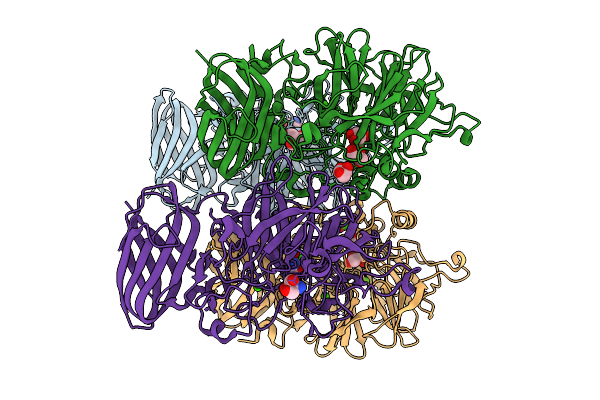 |
The Bifunctional Arabinofuranosidase/Xylosidase From Metagenome Of Pseudacanthotermes Militaris.
Organism: Pseudacanthotermes militaris
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: CA, TRS, GOL |
 |
Crystal Structure Of Dihydrodipicolinate Synthase From Mycobacterium Tuberculosis In Complex With Pyruvate
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: LYASE Ligands: PYR, GOL, MG, EDO, CL, PEG |
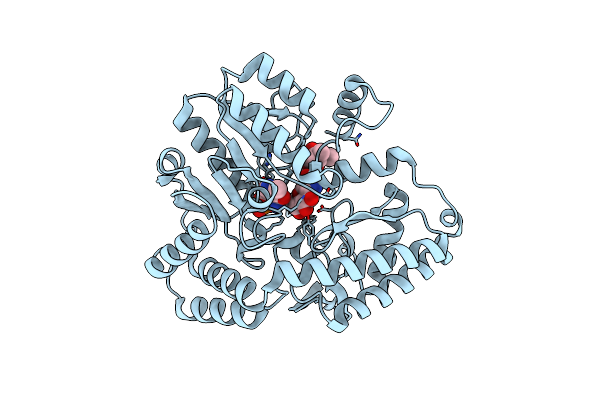 |
Organism: Micromonospora echinospora
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-09-11 Classification: ANTIBIOTIC Ligands: PMP, GET, CL |
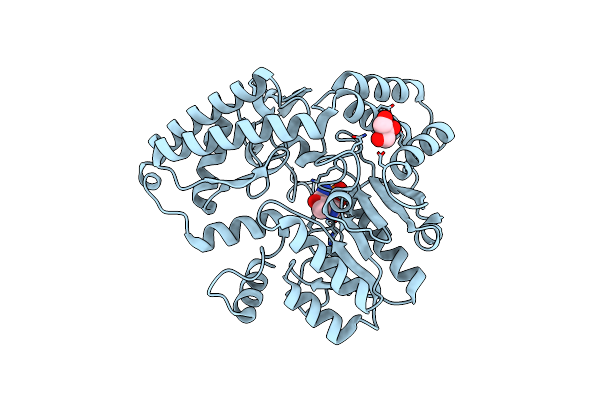 |
Organism: Micromonospora echinospora
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-09-11 Classification: ANTIBIOTIC Ligands: GOL, PMP, CL |
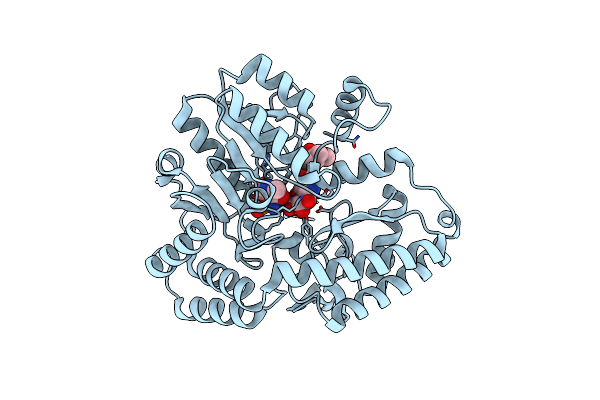 |
Organism: Micromonospora echinospora
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-09-11 Classification: BIOSYNTHETIC PROTEIN Ligands: 827, PMP, CL |
 |
Mycobacterium Tuberculosis Dihydrofolate Reductase In Complex With 5-(Cyclopropylethynyl)-6-(2-Fluorophenyl)Pyrimidine-2,4-Diamine
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-03-13 Classification: BIOSYNTHETIC PROTEIN Ligands: NDP, VB0, CO, SO4 |
 |
Mycobacterium Tuberculosis Dihydrofolate Reductase In Complex With N-(2-(2,6-Diamino-5-(Cyclopropylethynyl)Pyrimidin-4-Yl)Phenyl)Methanesulfonamide
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2024-03-13 Classification: BIOSYNTHETIC PROTEIN Ligands: NDP, PO4, CO, VBJ |
 |
Mycobacterium Tuberculosis Dihydrofolate Reductase In Complex With Methyl 4-(2,6-Diamino-5-(Cyclopropylethynyl)Pyrimidin-4-Yl)Benzoate
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-03-13 Classification: BIOSYNTHETIC PROTEIN Ligands: VFF, MES, NDP, CO, SO4 |
 |
Mycobacterium Tuberculosis Dihydrofolate Reductase In Complex With 5-(Cyclopropylethynyl)-6-(4-(Trifluoromethyl)Phenyl)Pyrimidine-2,4-Diamine
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-03-13 Classification: BIOSYNTHETIC PROTEIN Ligands: VFU, NDP, CO, SO4, MES |
 |
Mycobacterium Tuberculosis Dihydrofolate Reductase In Complex With N-(4-(2,6-Diamino-5-(Cyclopropylethynyl)Pyrimidin-4-Yl)Phenyl)Acetamide
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-13 Classification: BIOSYNTHETIC PROTEIN Ligands: VF6, NDP, CO, SO4, MES |
 |
Mycobacterium Tuberculosis Dihydrofolate Reductase In Complex With N-(4-(2,6-Diamino-5-(Cyclopropylethynyl)Pyrimidin-4-Yl)Phenyl)Methanesulfonamide
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-03-06 Classification: BIOSYNTHETIC PROTEIN Ligands: NDP, PO4, VCI, CO |
 |
Mycobacterium Tuberculosis Dihydrofolate Reductase In Complex With 5-(Cyclopropylethynyl)-6-(2-Methoxyphenyl)Pyrimidine-2,4-Diamine
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-03-06 Classification: BIOSYNTHETIC PROTEIN Ligands: NDP, VDT, CO, SO4 |
 |
Crystal Structure Of Candida Auris Dihydrofolate Reductase Complexed With Nadph And Cycloguanil
Organism: [candida] auris
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-08-02 Classification: HYDROLASE Ligands: VIL, NDP, SO4 |
 |
Organism: [candida] auris
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-06-14 Classification: HYDROLASE |
 |
Crystal Structure Of Candida Auris Dihydrofolate Reductase Complexed With Nadph
Organism: [candida] auris
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: NDP, EDO, NO3 |
 |
Crystal Structure Of Candida Auris Dihydrofolate Reductase Complexed With Nadph And Pyrimethamine
Organism: [candida] auris
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: NDP, CP6, NO3, EPE |
 |
Crystal Structure Of Mycobacterium Thermoresistibile Mure In Complex With Adp And 2,6-Diaminopimelic Acid
Organism: Mycolicibacterium thermoresistibile
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-04-05 Classification: LIGASE Ligands: MG, GOL, ADP, API |
 |
Crystal Structure Of Btrk, A Decarboxylase Involved In Butirosin Biosynthesis
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-08-17 Classification: LYASE Ligands: PLP, PEG |
 |
Crystal Structure Of Olmo, A Spirocyclase Involved In The Biosynthesis Of Oligomycin
Organism: Streptomyces avermitilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-08-17 Classification: HYDROLASE Ligands: MLT |
 |
Crystal Structure Of Osso, A Spirocyclase Involved In The Biosynthesis Of Ossamycin
Organism: Streptomyces ossamyceticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-08-17 Classification: HYDROLASE |

