Search Count: 23
 |
Crystal Structure Of Arabinose Isomerase From Hyper Thermophilic Bacterium Thermotoga Maritima (Tmai) Triple Mutant (K264A, E265A, K266A)
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:3.61 Å Release Date: 2021-10-20 Classification: ISOMERASE Ligands: MN |
 |
Organism: Alicyclobacillus sp. tp-7, Geobacillus kaustophilus (strain hta426), Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2021-10-20 Classification: ISOMERASE Ligands: MN, NA |
 |
Organism: Alicyclobacillus sp. tp-7, Geobacillus kaustophilus (strain hta426), Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2021-09-08 Classification: ISOMERASE Ligands: GOL, MN |
 |
Organism: Geobacillus kaustophilus (strain hta426), Alicyclobacillus sp. tp-7, Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-09-08 Classification: ISOMERASE Ligands: MPD, MN, RB0 |
 |
Crystal Structure Of Arabinose Isomerase From Hyper Thermophilic Bacterium Thermotoga Maritima (Tmai) Wt
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:3.53 Å Release Date: 2021-09-01 Classification: ISOMERASE Ligands: MN, GOL |
 |
Organism: Geobacillus kaustophilus (strain hta426), Alicyclobacillus sp. tp-7, Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-09-01 Classification: ISOMERASE Ligands: MPD, MN, GOL |
 |
Crystal Structure Of Pyrrolidone Carboxyl Peptidase From Thermophilic Keratin Degrading Bacterium Fervidobacterium Islandicum Aw-1 (Fipcp)
Organism: Fervidobacterium islandicum
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: GOL, 1PE |
 |
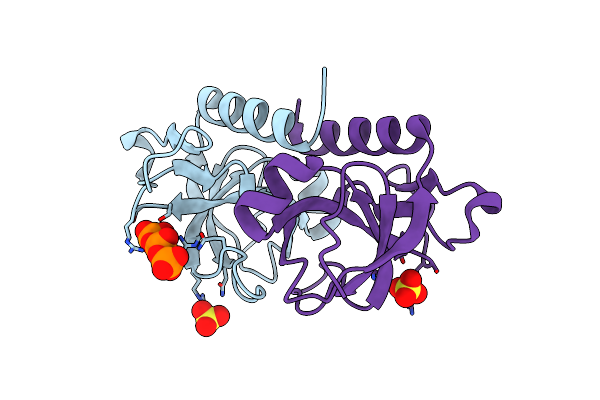
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-02-10 Classification: TOXIN Ligands: MES, GOL |
 |
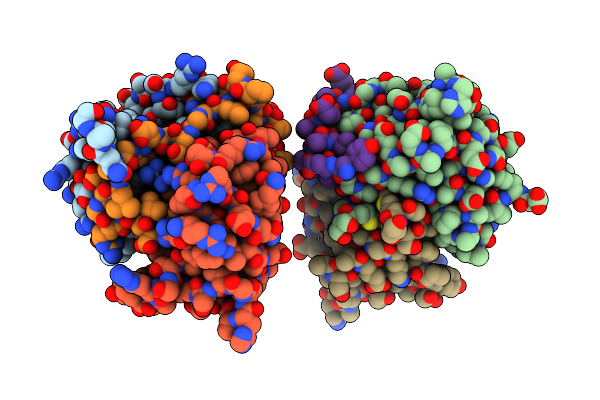
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-02-10 Classification: TOXIN Ligands: 3PO, SO4 |
 |
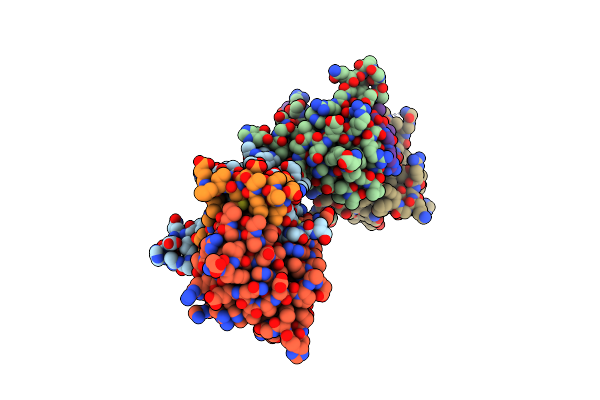
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-02-10 Classification: TOXIN |
 |
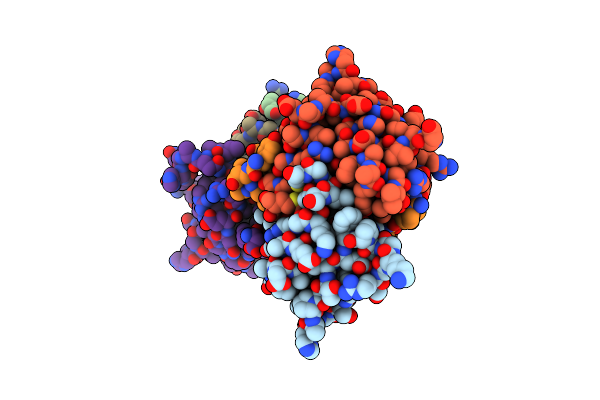
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2021-02-10 Classification: TOXIN |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2021-02-10 Classification: TOXIN |
 |
Crystal Structure Of Betaaspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum-Aw-1
Organism: Fervidobacterium islandicum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-12-30 Classification: METAL BINDING PROTEIN Ligands: GOL, PEG, ZN |
 |
Crystal Structure Of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 In Complex With Beta-Asp-Leu Dipeptide
Organism: Fervidobacterium islandicum
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2020-12-30 Classification: METAL BINDING PROTEIN Ligands: GOL, FWO, ZN, PEG |
 |
Crystal Structure Of Cytochrome Cl From The Marine Methylotrophic Bacterium Methylophaga Aminisulfidivorans Mpt (Ma-Cytcl)
Organism: Methylophaga aminisulfidivorans mp
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2020-07-22 Classification: ELECTRON TRANSPORT Ligands: HEC, EPE, GOL, CA, FE, NA |
 |
Crystal Structure Of Class A B-Lactamase, Penl, Variant Cys69Tyr, From Burkholderia Thailandensis
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-01-15 Classification: HYDROLASE |
 |
Crystal Structure Of Class A B-Lactamase, Penl, Variant Cys69Tyr, From Burkholderia Thailandensis, In Complex With Ceftazidime-Like Boronic Acid
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: CB4, GOL |
 |
Crystal Structure Of Class A B-Lactamase, Penl, Variant Asn136Asp, From Burkholderia Thailandensis
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-01-15 Classification: HYDROLASE |
 |
Crystal Structure Of Class A B-Lactamase, Penl, Variant Asn136Asp, From Burkholderia Thailandensis, In Complex With Ceftazidime-Like Boronic Acid
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: CB4, ACT |
 |
Crystal Structure Of Thermostable Cysteine Desulfurase (Fisufs) From Thermophilic Fervidobacterium Islandicum Aw-1
Organism: Fervidobacterium islandicum
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2019-10-09 Classification: TRANSFERASE Ligands: PLP, GOL, CIT, PEG |

