Search Count: 40
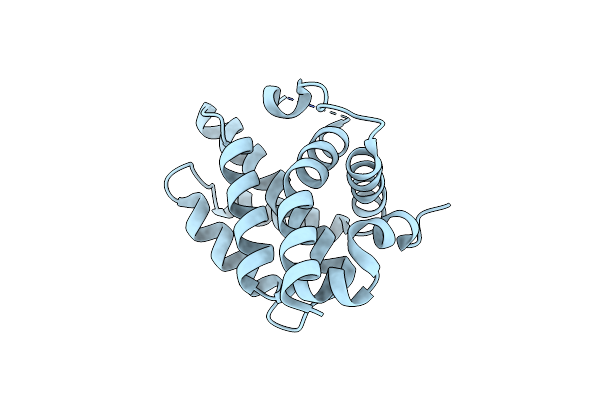 |
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-06-04 Classification: APOPTOSIS |
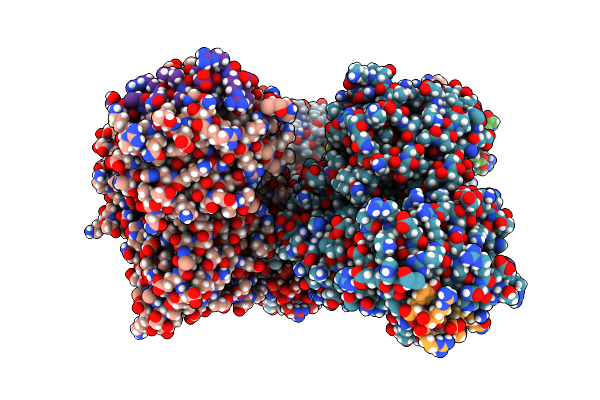 |
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-06-04 Classification: APOPTOSIS |
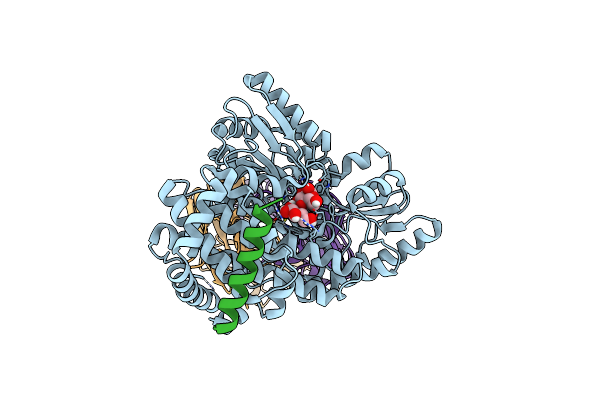 |
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: APOPTOSIS |
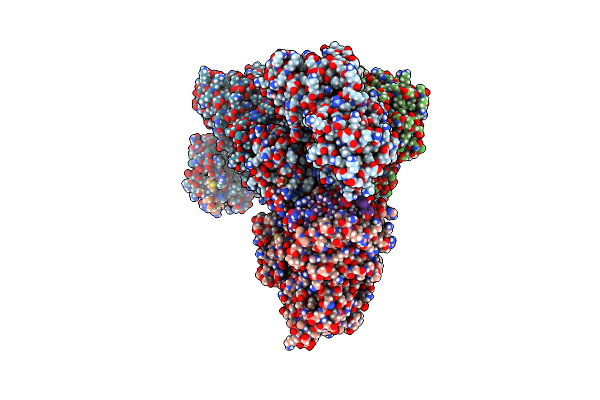 |
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-06-04 Classification: APOPTOSIS |
 |
Crystal Structure Of Peptidyl-Trna Hydrolase From Enterococcus Faecium At 1.22 A
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2025-02-19 Classification: HYDROLASE Ligands: EDO, BME, NA, SO4 |
 |
Crystal Structure Of Peptidyl-Trna Hydrolase Mutant From Enterococcus Faecium
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-04-03 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2024-02-07 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: VJX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2024-02-07 Classification: HYDROLASE Ligands: VXE |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2024-01-17 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.59 Å Release Date: 2023-11-29 Classification: TRANSCRIPTION |
 |
Organism: Human respiratory syncytial virus a (strain a2), Human respiratory syncytial virus a, Escherichia phage t2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.74 Å Release Date: 2020-11-11 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-11-11 Classification: IMMUNE SYSTEM |
 |
Structural Basis Of The Activation Of Heterotrimeric Gs-Protein By Isoproterenol-Bound Beta1-Adrenergic Receptor
Organism: Bos taurus, Lama glama, Meleagris gallopavo
Method: ELECTRON MICROSCOPY Release Date: 2020-09-02 Classification: SIGNALING PROTEIN Ligands: 5FW |
 |
L-Methionine Depletion With An Engineered Human Enzyme Disrupts Prostate Cancer Metabolism
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2020-04-22 Classification: LYASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2018-09-19 Classification: TRANSFERASE Ligands: SAH, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2018-09-19 Classification: TRANSFERASE Ligands: SAH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-09-19 Classification: TRANSFERASE |
 |
Structural Insights Into The Induced-Fit Inhibition Of Fascin By A Small Molecule
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-04-18 Classification: PROTEIN BINDING Ligands: C7V |
 |
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-12-23 Classification: HYDROLASE |
 |
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-12-23 Classification: HYDROLASE Ligands: GD |

