Search Count: 30
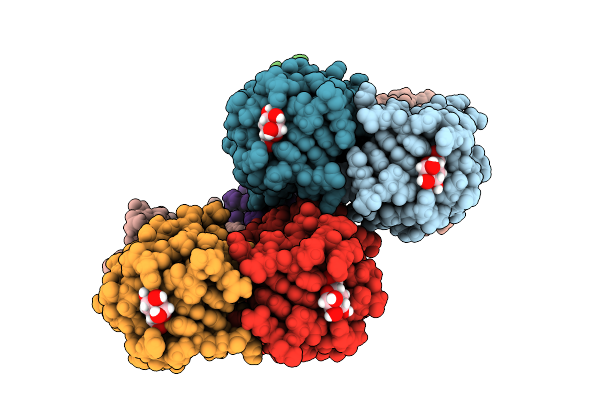 |
Joint X-Ray/Neutron Room Temperature Structure Of Perdeuterated Leca Lectin In Complex With Deuterated Galactose
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-12-24 Classification: SUGAR BINDING PROTEIN Ligands: GLA, CA |
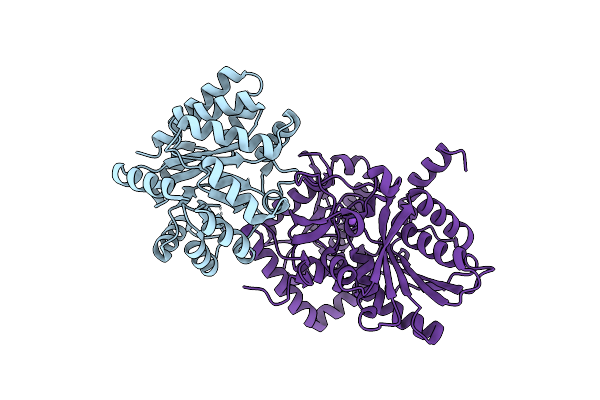 |
Joint X-Ray/Neutron Structure Of Salmonella Typhimurium Tryptophan Synthase Internal Aldimine From Microgravity-Grown Crystal
Organism: Salmonella enterica subsp. enterica serovar typhi, Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.80 Å, 2.10 Å Release Date: 2024-02-14 Classification: LYASE |
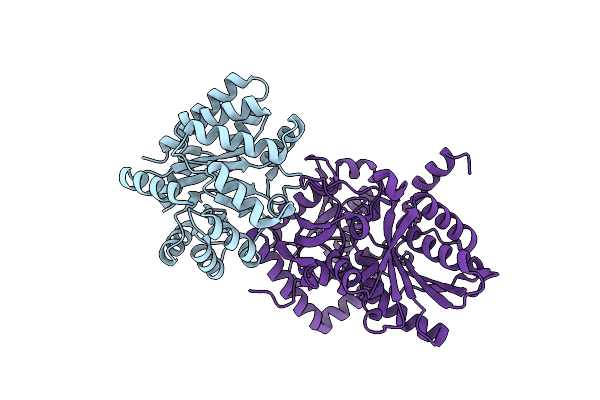 |
X-Ray Crystal Structure Of Salmonella Typhimurium Tryptophan Synthase Internal Aldimine At Ph 5.0
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-02-14 Classification: LYASE |
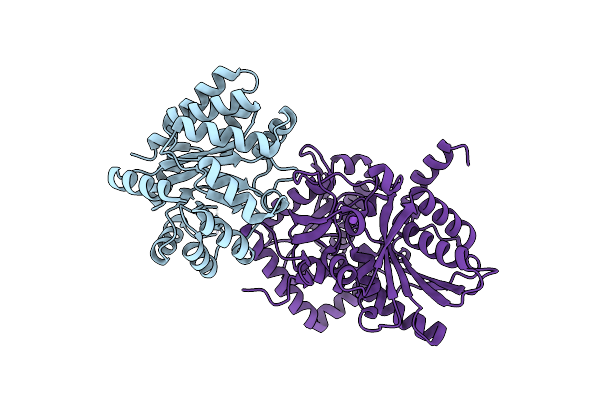 |
X-Ray Crystal Structure Of Salmonella Typhimurium Tryptophan Synthase Internal Aldimine
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-02-14 Classification: LYASE |
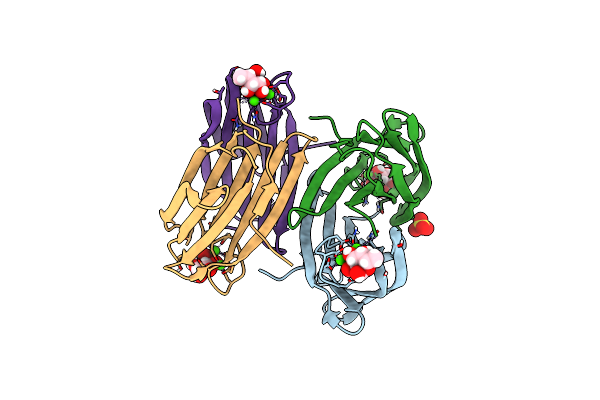 |
Joint X-Ray/Neutron Room Temperature Structure Of Perdeuterated Lecb Lectin In Complex With Perdeuterated Fucose
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.85 Å, 1.90 Å Release Date: 2022-01-12 Classification: SUGAR BINDING PROTEIN Ligands: CA, FUC, SO4 |
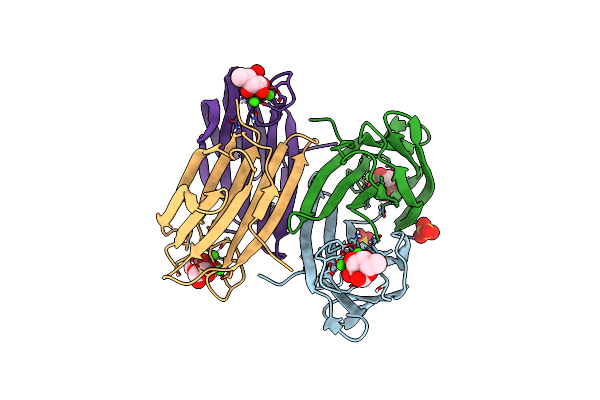 |
X-Ray Crystal Structure Of Perdeuterated Lecb Lectin In Complex With Perdeuterated Fucose
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:0.90 Å Release Date: 2022-01-12 Classification: SUGAR BINDING PROTEIN Ligands: FUC, CA, SO4 |
 |
X-Ray Crystal Structure Of Aspartate Alpha-Decarboxylase In Complex With D-Serine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-10-06 Classification: LYASE Ligands: DSN, EDO, SER |
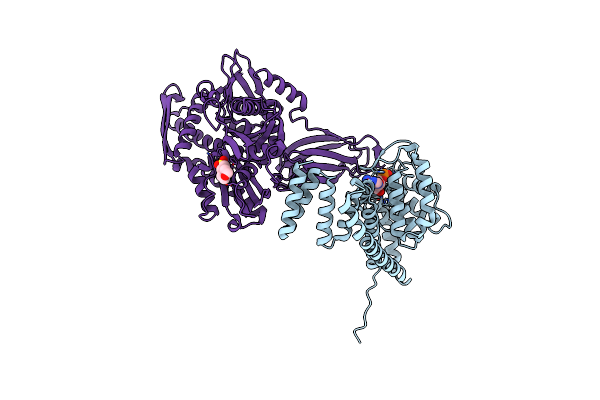 |
Organism: Cricetulus griseus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-07-07 Classification: HYDROLASE Ligands: AMP, MES, MG |
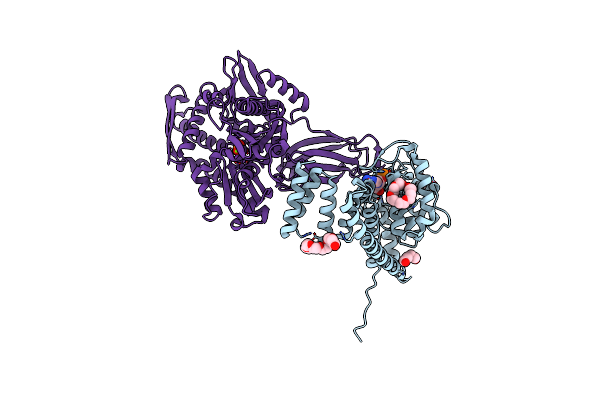 |
Organism: Cricetulus griseus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2021-07-07 Classification: HYDROLASE Ligands: AMP, MG, PO4, K, P33, PEG |
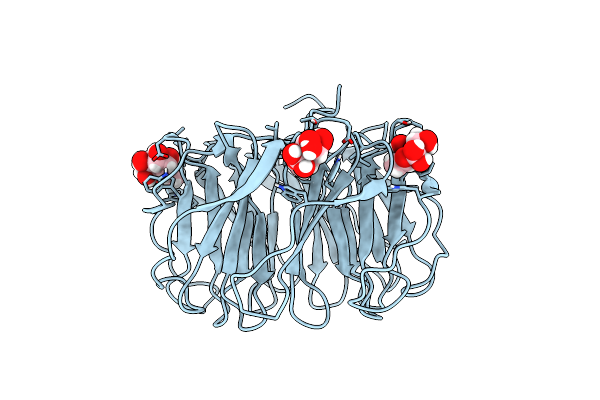 |
Joint X-Ray/Neutron Room Temperature Structure Of Perdeuterated Pll Lectin In Complex With Perdeuterated L-Fucose
Organism: Photorhabdus laumondii
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.84 Å, 2.2 Å Release Date: 2021-03-24 Classification: SUGAR BINDING PROTEIN Ligands: FUL, FUC |
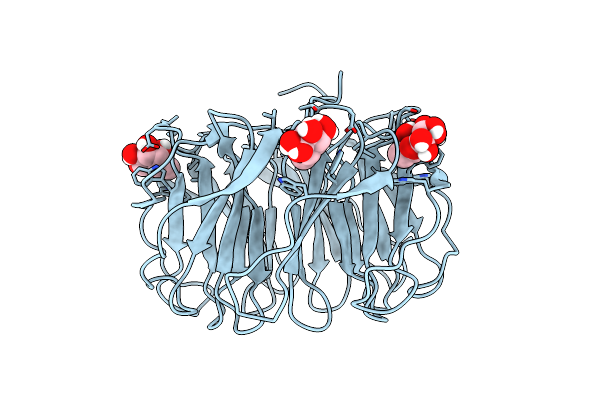 |
Room Temperature X-Ray Structure Of Perdeuterated Pll Lectin In Complex With L-Fucose
Organism: Photorhabdus laumondii
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2021-03-17 Classification: SUGAR BINDING PROTEIN Ligands: FUL, FUC |
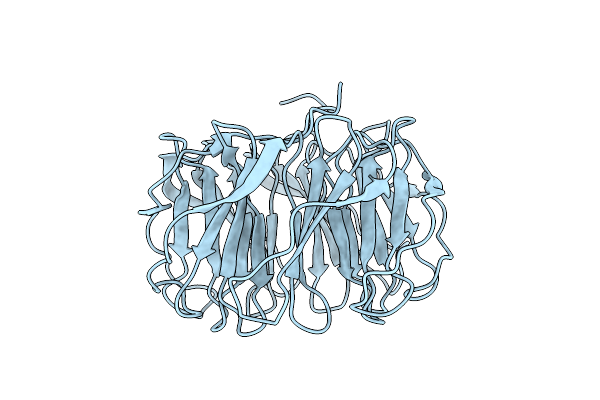 |
Organism: Photorhabdus laumondii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-03-17 Classification: SUGAR BINDING PROTEIN |
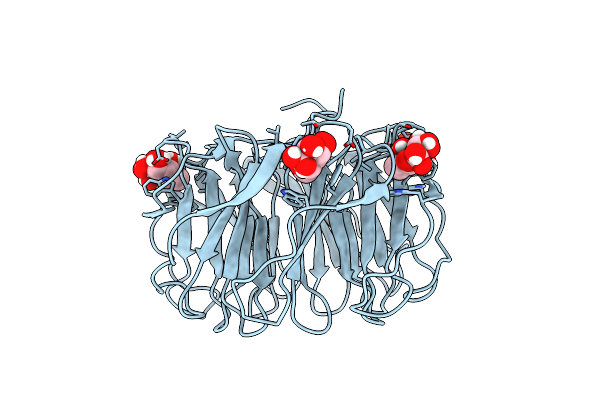 |
Room Temperature X-Ray Structure Of H/D-Exchanged Pll Lectin In Complex With L-Fucose
Organism: Photorhabdus laumondii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-03-17 Classification: SUGAR BINDING PROTEIN Ligands: FUC, FUL |
 |
Organism: Photorhabdus laumondii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-03-17 Classification: SUGAR BINDING PROTEIN Ligands: FUL, FUC, GOL |
 |
Organism: Photorhabdus laumondii
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.70 Å, 2.20 Å, Release Date: 2021-03-17 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.9000 Å, 2.2220 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, DOD |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.9000 Å, 2.0900 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: HEM, K, SO4, ASC, DOD |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-01-24 Classification: TRANSCRIPTION Ligands: PO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2018-01-24 Classification: TRANSCRIPTION |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.806 Å, 2.202 Å Release Date: 2016-12-21 Classification: OXIDOREDUCTASE Ligands: HEM, K, SO4, DOD |

