Search Count: 21
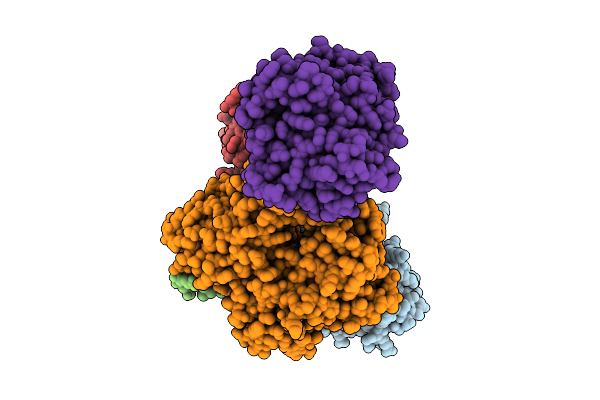 |
Viral Protein Dp71L In Complex With Phosphorylated Eif2Alpha (Ntd) And Protein Phosphatase 1A (D64A), Stabilized By G-Actin/Dnasei
Organism: African swine fever virus, Homo sapiens, Oryctolagus cuniculus, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: TRANSLATION Ligands: MN, CA, ATP |
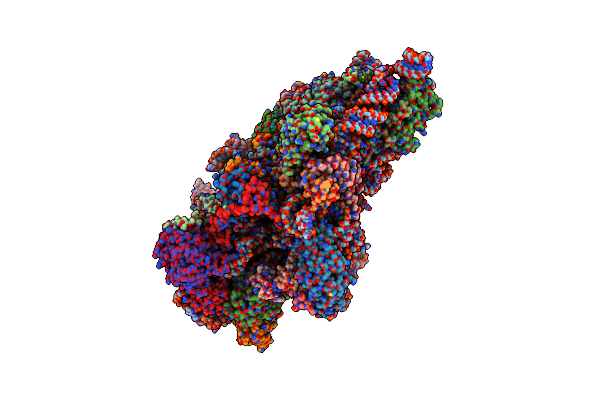 |
Cryoem Structure Of The Human 40S Small Ribosomal Subunit In Complex With Translation Initiation Factors Eif1A And Eif5B.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2022-04-27 Classification: RIBOSOME Ligands: GNP, 5GP, ZN |
 |
Method: ELECTRON MICROSCOPY
Release Date: 2020-09-23 Classification: RIBOSOME Ligands: ZN, GDP, U6A, MET |
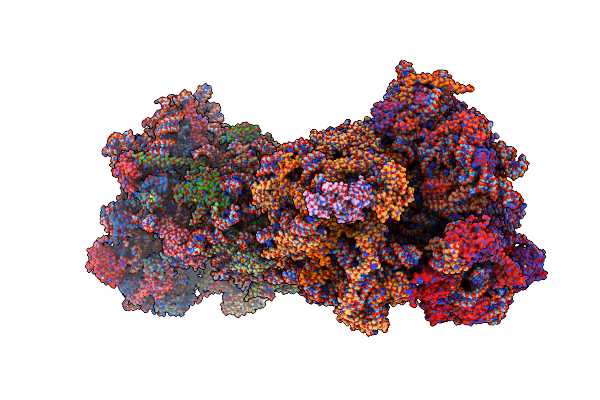 |
Coping With Proline Stalling: Structural Basis Of Hypusine-Induced Protein Synthesis By The Eukaryotic Ribosome
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2017-01-25 Classification: RIBOSOME Ligands: MG, OHX, ZN, SPS, PRO |
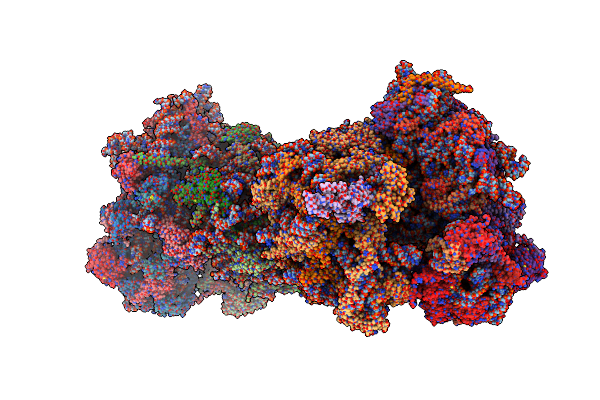 |
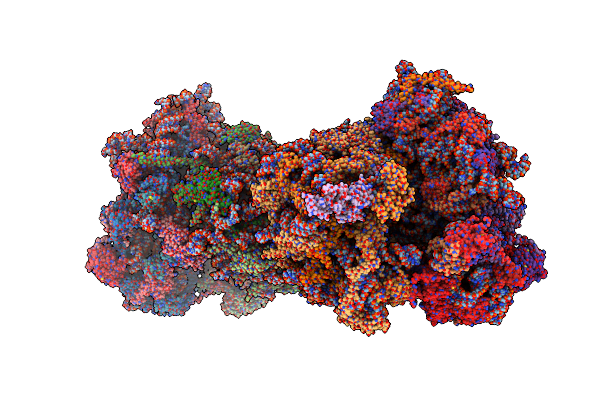
Crystal Structure Of The S.Cerevisiae 80S Ribosome In Complex With The A-Site Bound Aminoacyl-Trna Analog Acca-Pro
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2017-01-18 Classification: RIBOSOME Ligands: OHX, MG, ZN, PHE, LEU, SPS, 8AN |
 |
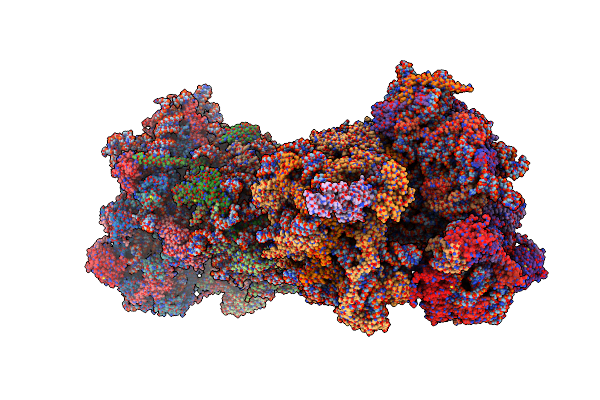
Complex Of Yeast 80S Ribosome With Hypusine-Containing/Non-Modified Eif5A And/Or A Peptidyl-Trna Analog
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2016-12-14 Classification: RIBOSOME Ligands: ZN, SPS, MG, OHX |
 |
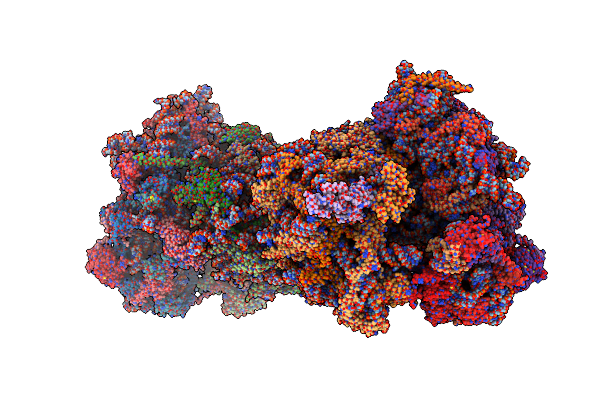
Complex Of Yeast 80S Ribosome With Hypusine-Containing/Non-Modified Eif5A And/Or A Peptidyl-Trna Analog
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-12-14 Classification: RIBOSOME Ligands: ZN, SPS, MG |
 |
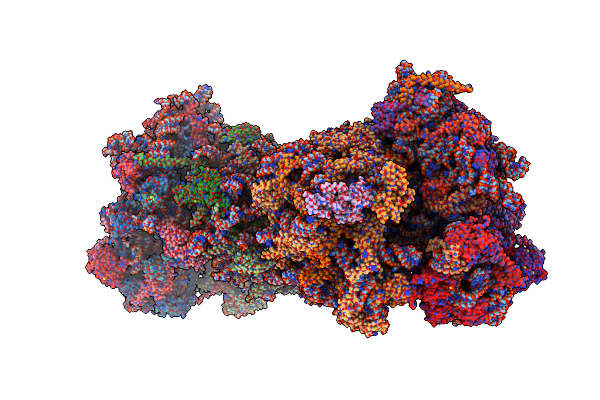
Crystal Structure Of The S.Cerevisiae 80S Ribosome In Complex With The A-Site Bound Aminoacyl-Trna Analog Accpmn
Organism: Synthetic construct, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2016-11-23 Classification: RIBOSOME Ligands: MG, OHX, ZN |
 |
Crystal Structure Of The S.Cerevisiae 80S Ribosome In Complex With The A-Site Bound Aminoacyl-Trna Analog Acca-Pro
Organism: Synthetic construct, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2016-11-23 Classification: RIBOSOME Ligands: OHX, MG, ZN, PRO |
 |
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2016-08-31 Classification: RIBOSOME Ligands: MG, OHX, ZN |
 |
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2016-06-01 Classification: RIBOSOME Ligands: ZN |
 |
Structure Of The Kluyveromyces Lactis 80S Ribosome In Complex With The Cricket Paralysis Virus Ires And Eef2
Organism: Cricket paralysis virus, Saccharomyces cerevisiae p283, Kluyveromyces lactis, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2016-05-18 Classification: RIBOSOME Ligands: MG, ZN, GCP, 6EM |
 |
Structure Of The Yeast Kluyveromyces Lactis Small Ribosomal Subunit In Complex With The Cricket Paralysis Virus Ires.
Organism: Cricket paralysis virus, Kluyveromyces lactis
Method: ELECTRON MICROSCOPY Resolution:3.80 Å Release Date: 2016-05-18 Classification: RIBOSOME Ligands: MG, ZN |
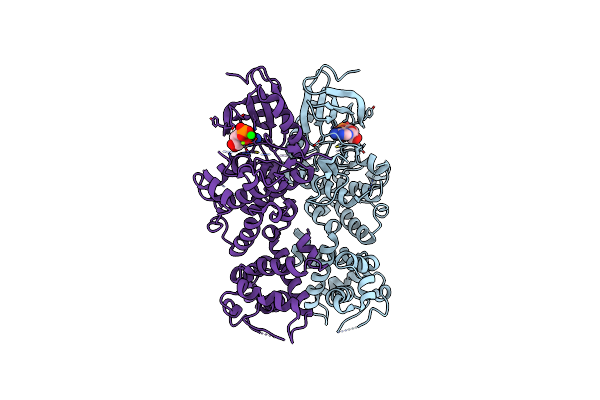 |
Structure Of The Dual Enzyme Ire1 Reveals The Basis For Catalysis And Regulation Of Non-Conventional Splicing
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-01-29 Classification: HYDROLASE, TRANSFERASE Ligands: MG, SR, ADP |
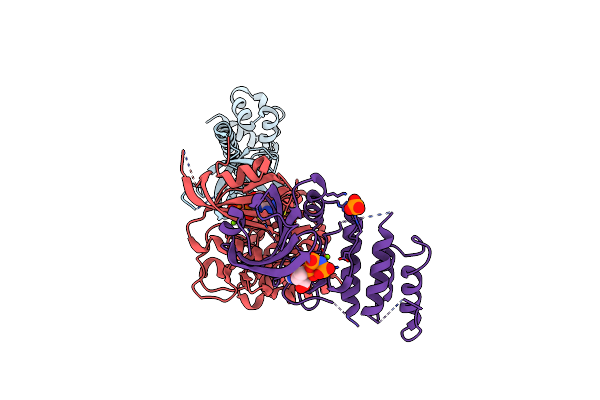 |
Organism: Saccharomyces cerevisiae, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2005-09-27 Classification: protein synthesis/transferase Ligands: MG, ANP, PO4 |
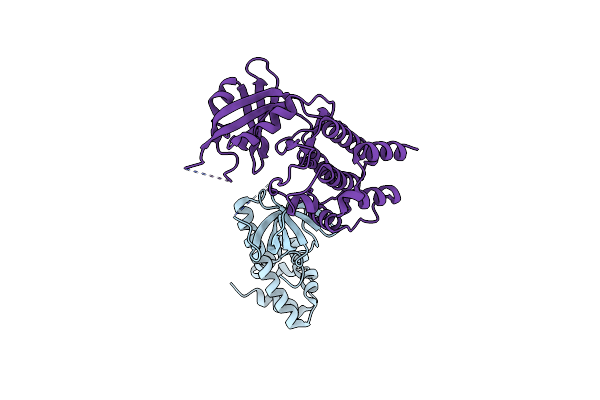 |
Organism: Saccharomyces cerevisiae, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-09-27 Classification: protein synthesis/transferase |
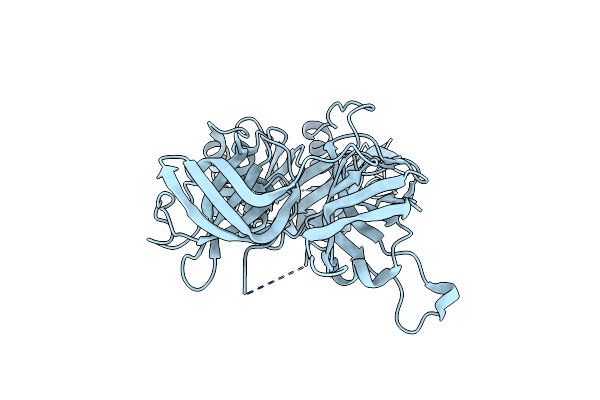 |
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2004-01-20 Classification: TRANSLATION Ligands: ZN |
 |
Organism: Myxoma virus (strain lausanne)
Method: SOLUTION NMR Release Date: 2002-03-06 Classification: VIRAL PROTEIN |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2000-12-06 Classification: TRANSLATION |
 |
X-Ray Structure Of Translation Initiation Factor If2/Eif5B Complexed With Gdp
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-12-06 Classification: TRANSLATION Ligands: GDP |

