Search Count: 29
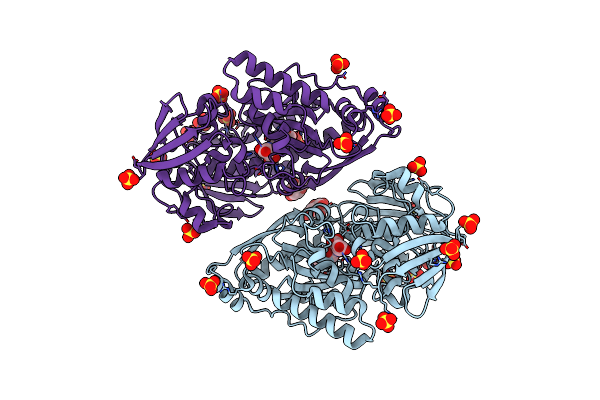 |
Structure In P3212 Form Of The Pbp/Sbp Moaa In Complex With Mannopinic Acid From A.Tumefacien R10
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-01-22 Classification: TRANSPORT PROTEIN Ligands: GLC, N72, SO4, CL |
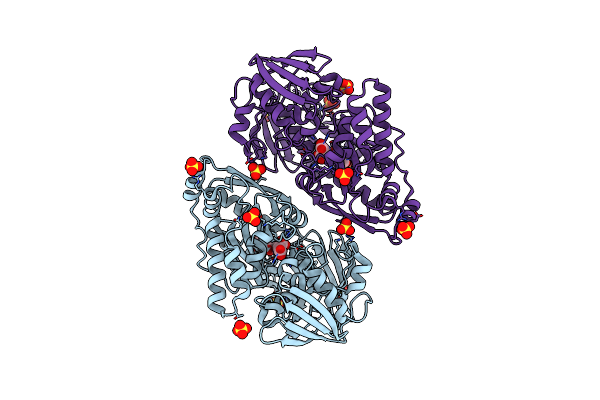 |
Structure In P3212 Form Of The Pbp/Sbp Moaa In Complex With Glucopinic Acid From A.Tumefacien R10
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-01-22 Classification: TRANSPORT PROTEIN Ligands: N7T, SO4, CL |
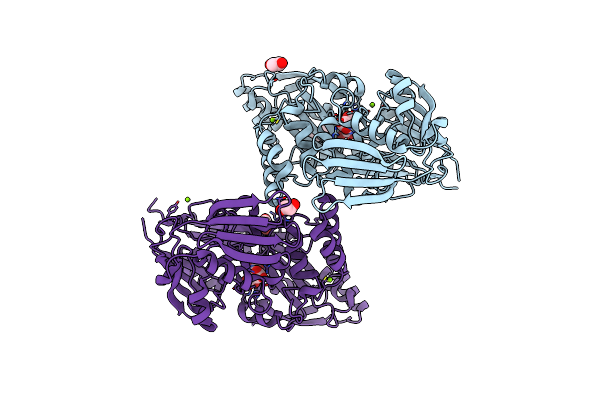 |
Structure In P21 Form Of The Pbp/Sbp Moaa In Complex With Mannopinic Acid From A.Tumefacien R10
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2020-01-22 Classification: TRANSPORT PROTEIN Ligands: N72, EDO, PEG, MG |
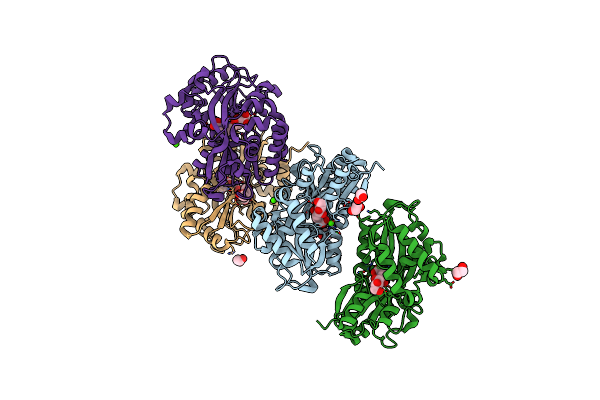 |
Structure Of The Pbp/Sbp Mota In Complex With Mannopinic Acid From A.Tumefacien R10
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2020-01-22 Classification: TRANSPORT PROTEIN Ligands: N72, EDO, CA, PEG |
 |
Crystal Structure Of The Pbp/Sbp Mota In Complex With Glucopinic Acid From A. Tumefaciens B6/R10
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-01-22 Classification: TRANSPORT PROTEIN Ligands: N7T, EDO |
 |
Structure Of The Pbp Agaa In Complex With Agropinic Acid From A.Tumefacien R10
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-12-26 Classification: TRANSPORT PROTEIN Ligands: GOL, G9Z, MES, ACT, ZN, SO4, EDO, PEG |
 |
Structure In P212121 Form Of The Pbp Agtb In Complex With Agropinic Acid From A.Tumefacien R10
Organism: Agrobacterium tumefaciens lba4213 (ach5)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2018-12-26 Classification: TRANSPORT PROTEIN Ligands: EDO, G9Z |
 |
Structure In C2 Form Of The Pbp Agtb From A.Tumefacien R10 In Complex With Agropinic Acid
Organism: Agrobacterium tumefaciens lba4213 (ach5)
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2018-12-26 Classification: TRANSPORT PROTEIN Ligands: EDO, P4G, G9Z, PEG, P6G |
 |
Structure In P1 Form Of The Pbp Agtb In Complex With Agropinic Acid From A.Tumefacien R10
Organism: Agrobacterium tumefaciens lba4213 (ach5)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2018-12-26 Classification: TRANSPORT PROTEIN Ligands: G9Z, EDO, NA |
 |
Structure Of Agrobacterium Tumefaciens B6 Strain Pbp Soca Complexed With Deoxyfructosylglutamine (Dfg) At 1.8 A Resolution
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-10-31 Classification: TRANSPORT PROTEIN Ligands: SNW, EDO |
 |
Organism: Agrobacterium
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-10-10 Classification: TRANSPORT PROTEIN Ligands: 2W2, PEG, EDO, NA |
 |
Structure Of The Periplasmic Binding Protein (Pbp) Occj From Agrobacterium Tumefaciens B6
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-12-20 Classification: DNA Ligands: EDO, CL |
 |
Structure Of The Periplasmic Binding Protein (Pbp) Occj From A. Tumefaciens B6 In Complex With Octopine.
Organism: Agrobacterium tumefaciens str. b6
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2017-12-20 Classification: Octopine-binding protein Ligands: EDO, NA, CL, GOL, 6DB, ACT |
 |
Structure Of The Periplasmic Binding Protein (Pbp) Noct-G97S Mutant From A. Tumefaciens C58 In Complex With Octopine.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-12-20 Classification: PROTEIN BINDING Ligands: 6DB, EDO, SO4, CL, PEG |
 |
Structure Of The Periplasmic Binding Protein (Pbp) Noct From A.Tumefaciens C58 In Complex With Histopine.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2017-12-20 Classification: PROTEIN BINDING Ligands: EDO, AOZ |
 |
Structure Of The Periplasmic Binding Protein (Pbp) Noct From Agrobacterium Tumefaciens C58 In Complex With Octopinic Acid
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-12-20 Classification: PROTEIN BINDING Ligands: AQQ, EDO |
 |
Structure Of The Periplasmic Binding Protein (Pbp) Noct From Agrobacterium Tumefaciens C58 In Complex With Noroctopinic Acid.
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-12-20 Classification: PROTEIN BINDING Ligands: AQK, EDO, CL, PEG, PGE |
 |
Structure Of The Periplasmic Binding Protein M117N-Noct From A. Tumefaciens In Complex With Octopine
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2016-11-30 Classification: MEMBRANE PROTEIN Ligands: PEG, EDO, 1PE, 6DB, TOE |
 |
Structure Of The Periplasmic Binding Protein Noct From A.Tumefaciens In Complex With Octopine
Organism: Agrobacterium fabrum
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-11-30 Classification: MEMBRANE PROTEIN Ligands: 6DB, EDO, PEG |
 |
Crystal Structure Of The Pbp Mota In Complex With Mannopine From A. Tumefaciens B6
Organism: Agrobacterium tumefaciens str. b6
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-09-21 Classification: TRANSPORT PROTEIN Ligands: MO0, EDO, CA |

