Search Count: 17
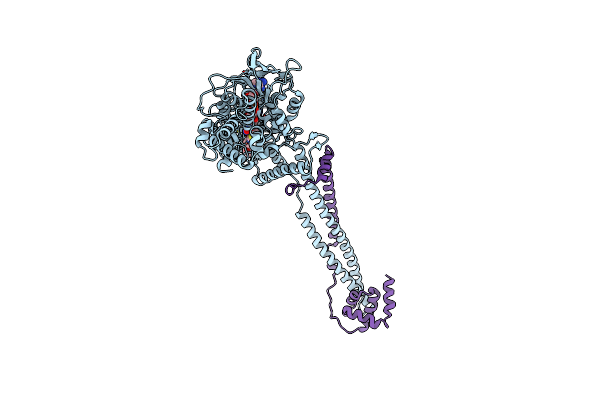 |
Thieno[3,2-B]Pyrrole-5-Carboxamides As Novel Reversible Inhibitors Of Histone Lysine Demethylase Kdm1A/Lsd1: Compound 19
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2017-02-22 Classification: OXIDOREDUCTASE Ligands: FAD, 6W0 |
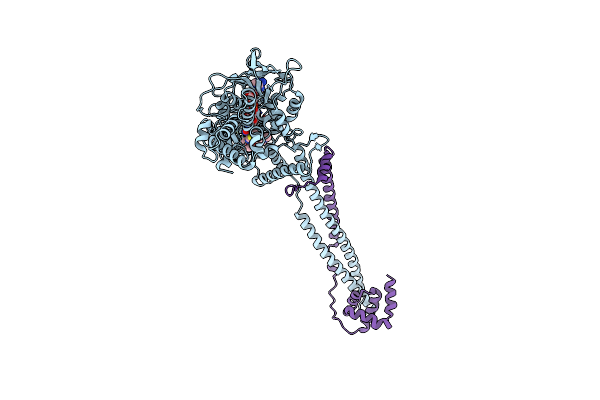 |
Thieno[3,2-B]Pyrrole-5-Carboxamides As Novel Reversible Inhibitors Of Histone Lysine Demethylase Kdm1A/Lsd1: Compound 15
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-02-22 Classification: OXIDOREDUCTASE Ligands: FAD, 6W3 |
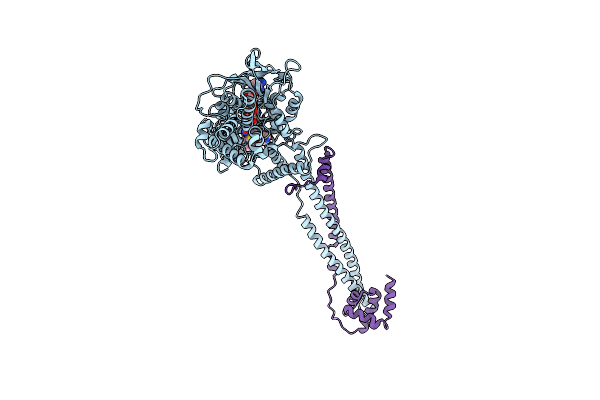 |
Thieno[3,2-B]Pyrrole-5-Carboxamides As Novel Reversible Inhibitors Of Histone Lysine Demethylase Kdm1A/Lsd1: Compound 34
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2017-02-22 Classification: OXIDOREDUCTASE Ligands: FAD, 6W1 |
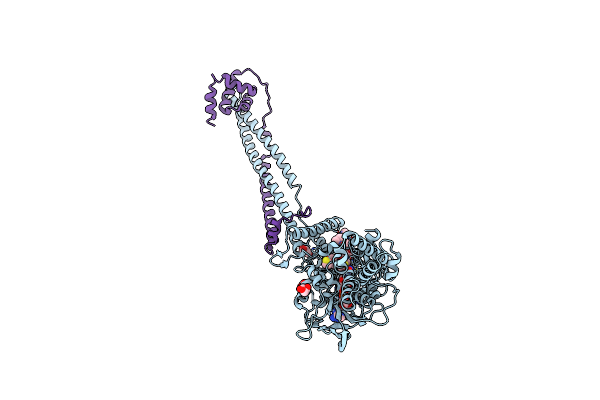 |
Structure Of The Kdm1A/Corest Complex With The Inhibitor 4-Methyl-N-[4-[[4-(1-Methylpiperidin-4-Yl)Oxyphenoxy]Methyl]Phenyl]Thieno[3,2-B]Pyrrole-5-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2017-02-22 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, 6X3 |
 |
Structure Of The Kdm1A/Corest Complex With The Inhibitor 4-Ethyl-N-[3-(Methoxymethyl)-2-[[4-[[(3R)-Pyrrolidin-3-Yl]Methoxy]Phenoxy]Methyl]Phenyl]Thieno[3,2-B]Pyrrole-5-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2017-02-22 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, 6X0 |
 |
Structure Of The Kdm1A/Corest Complex With The Inhibitor N-[3-(Ethoxymethyl)-2-[[4-[[(3R)-Pyrrolidin-3-Yl]Methoxy]Phenoxy]Methyl]Phenyl]-4-Methylthieno[3,2-B]Pyrrole-5-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2017-02-22 Classification: OXIDOREDUCTASE Ligands: FAD, 6X5, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE Ligands: FAD, M8A |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-09-10 Classification: TRANSCRIPTION Ligands: D69 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-09-10 Classification: TRANSCRIPTION Ligands: D70 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2014-09-10 Classification: TRANSCRIPTION Ligands: D73 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-09-10 Classification: TRANSCRIPTION Ligands: D51 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2014-09-10 Classification: TRANSCRIPTION Ligands: D52 |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2012-02-29 Classification: TRANSFERASE Ligands: SO4, GOL |
 |
Crystal Structure Of Adenosine Phosphorylase From Bacillus Cereus Complexed With Adenosine
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2012-02-29 Classification: TRANSFERASE Ligands: ADN, SO4, GOL |
 |
Crystal Structure Of Adenosine Phosphorylase From Bacillus Cereus Complexed With Inosine
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2012-02-29 Classification: TRANSFERASE Ligands: NOS, SO4, GOL |
 |
Crystal Structure Of Bacillus Cereus Adenosine Phosphorylase D204N Mutant Complexed With Adenosine
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2012-02-29 Classification: TRANSFERASE Ligands: ADN, SO4, GOL |
 |
Crystal Structure Of Bacillus Cereus Adenosine Phosphorylase D204N Mutant Complexed With Inosine
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2012-02-29 Classification: TRANSFERASE Ligands: NOS, SO4, GOL |

