Search Count: 13
 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-10-31 Classification: HYDROLASE Ligands: GOL, CL |
 |
Crystal Structure Of Penicillin-Binding Protein 4 (Pbp4) From Enterococcus Faecalis In The Benzylpenicillin Bound Form.
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2018-10-31 Classification: HYDROLASE Ligands: GOL, CL, PEG, PNM |
 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) From Enterococcus Faecium In The Open Conformation
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-10-31 Classification: PROTEIN BINDING Ligands: SO4 |
 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) From Enterococcus Faecium In The Imipenem-Bound Form
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-10-31 Classification: protein binding/antibiotic Ligands: IM2, SO4 |
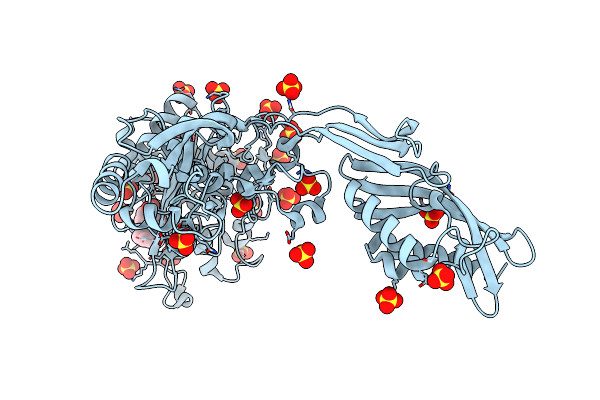 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) From Enterococcus Faecium In The Benzylpenicilin-Bound Form
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2018-10-31 Classification: protein binding/antibiotic Ligands: PNM, SO4 |
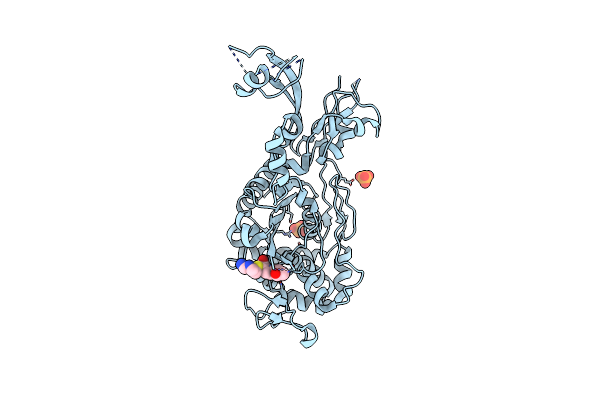 |
Crystal Structure Of Pencillin Binding Protein 4 (Pbp4) From Enterococcus Faecalis In The Imipenem-Bound Form
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2018-10-31 Classification: protein binding/antibiotic Ligands: IM2, PO4 |
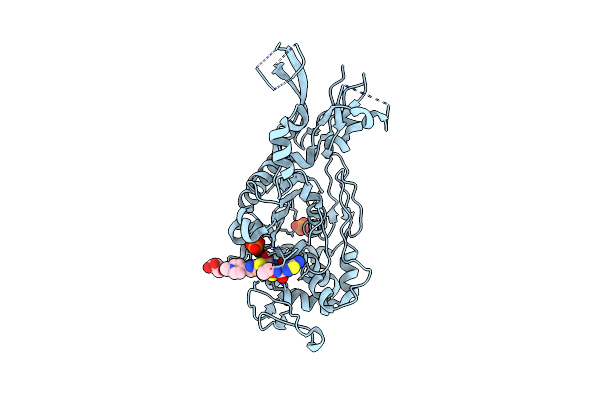 |
Crystal Structure Of Penicillin-Binding Protein 4 (Pbp4) From Enterococcus Faecalis In The Ceftaroline-Bound Form
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2018-10-31 Classification: protein binding/antibiotic Ligands: AI8, PO4, GOL |
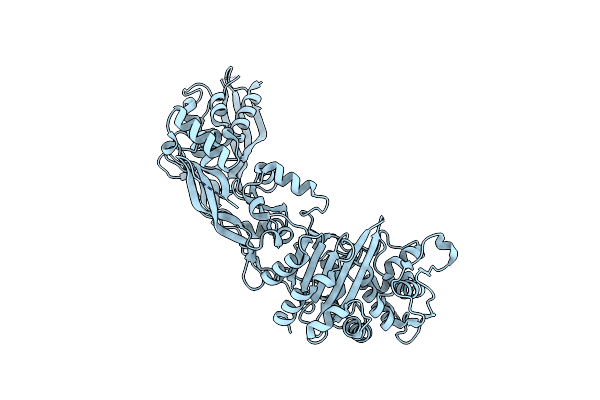 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) From Enterococcus Faecium In The Closed Conformation
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2018-10-31 Classification: PROTEIN BINDING |
 |
Udp-3-O-[3-Hydroxymyristoyl] N-Acetylglucosamine Deacetylase In Complex With Inhibitor
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2012-02-01 Classification: Hydrolase/Hydrolase inhibitor Ligands: ZN, RFN, GAI, EDO |
 |
Structure Guided Development Of Novel Thymidine Mimetics Targeting Pseudomonas Aeruginosa Thymidylate Kinase: From Hit To Lead Generation
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2012-02-01 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 0DF, MG |
 |
Structure Guided Development Of Novel Thymidine Mimetics Targeting Pseudomonas Aeruginosa Thymidylate Kinase: From Hit To Lead Generation
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-02-01 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 0DJ |
 |
Structure Guided Development Of Novel Thymidine Mimetics Targeting Pseudomonas Aeruginosa Thymidylate Kinase: From Hit To Lead Generation
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2012-02-01 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MG, 0DN |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-01-04 Classification: Hydrolase/Hydrolase inhibitor Ligands: ZN, 03I |

