Search Count: 29
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-01-12 Classification: DNA Ligands: 5N0, ACT |
 |
Rna Polymerase Ii Elongation Complex With Hairpin Polyamide Py-Im 1, Scaffold 1
Organism: Synthetic construct, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-01-12 Classification: DNA BINDING PROTEIN/DNA/RNA Ligands: 5N0, ZN, MG |
 |
Rna Polymerase Ii Elongation Complex With Hairpin Polyamide Py-Im 1, Scaffold 1 Soaked With Ctp
Organism: Synthetic construct, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2022-01-12 Classification: DNA BINDING PROTEIN/DNA/RNA Ligands: MG, 5N0, ZN, PPV |
 |
Organism: Synthetic construct, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-01-12 Classification: DNA BINDING PROTEIN/DNA/RNA Ligands: MG, ZN |
 |
Organism: Synthetic construct, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-01-12 Classification: DNA BINDING PROTEIN/DNA/RNA Ligands: MG, ZN |
 |
Rna Polymerase Ii Elongation Complex With Hairpin Polyamide Py-Im 1, Scaffold 2
Organism: Synthetic construct, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2022-01-12 Classification: DNA BINDING PROTEIN/DNA/RNA Ligands: MG, 5N0, ZN |
 |
Rna Polymerase Ii Elongation Complex With Hairpin Polyamide Py-Im 1, Scaffold 2 Soaked With Utp
Organism: Synthetic construct, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2022-01-12 Classification: DNA BINDING PROTEIN/DNA/RNA Ligands: MG, 5N0, ZN |
 |
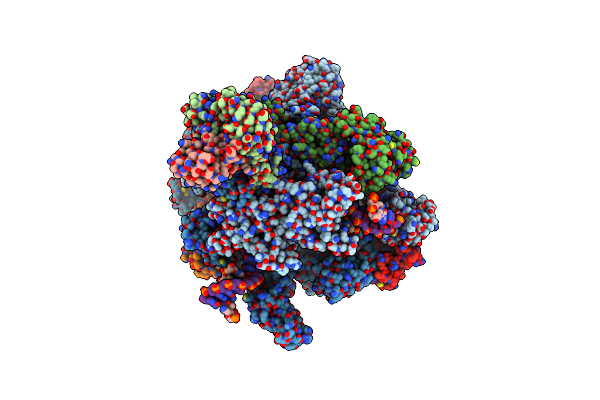
Organism: Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2017-11-22 Classification: TRANSCRIPTION/RNA/DNA Ligands: ZN, MG |
 |
Organism: Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2017-11-22 Classification: TRANSCRIPTION/RNA/DNA Ligands: ZN, MG |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2013-01-30 Classification: Hydrolase/DNA Ligands: NI, CA, CL |
 |
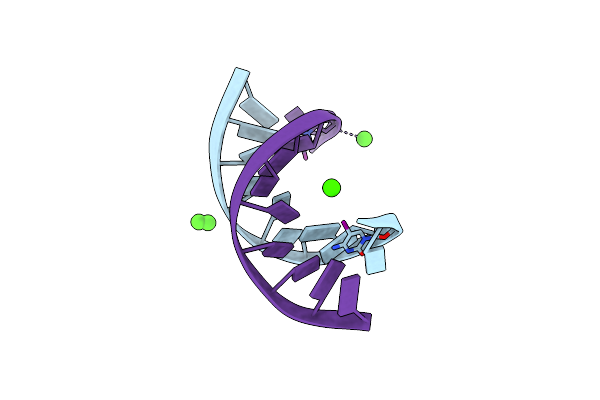
Crystal Structure Of The C-Terminal Domain Of Nicking Enzyme From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2013-01-30 Classification: HYDROLASE Ligands: CA |
 |
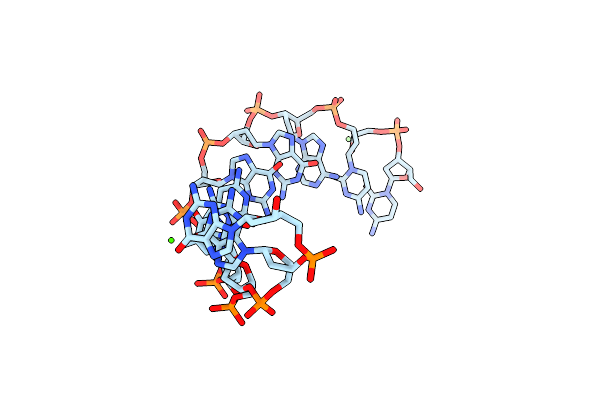
Structural Basis For Cyclic Py-Im Polyamide Allosteric Inhibition Of Nuclear Receptor Binding
Method: X-RAY DIFFRACTION
Resolution:0.95 Å Release Date: 2010-09-08 Classification: DNA Ligands: CA, 1P2 |
 |
Method: X-RAY DIFFRACTION
Resolution:0.98 Å Release Date: 2009-07-28 Classification: DNA Ligands: CA |
 |
Method: X-RAY DIFFRACTION
Resolution:1.18 Å Release Date: 2009-07-28 Classification: DNA Ligands: CA, 1P1 |
 |
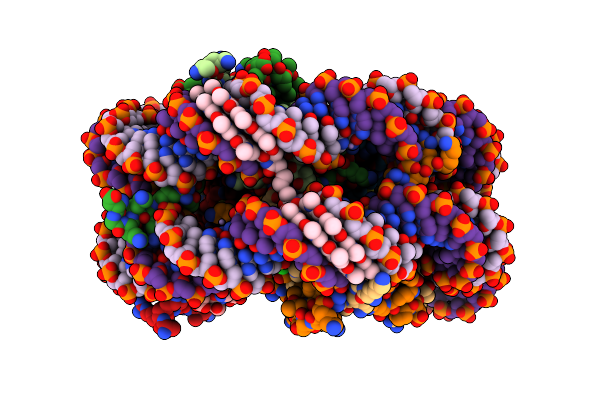 |
Organism: Xenopus laevis, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2004-05-11 Classification: STRUCTURAL PROTEIN/DNA Ligands: MN, IMT, PYB, ABU, BAL, DIB, OGG, CL |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2003-02-18 Classification: STRUCTURAL PROTEIN/DNA Ligands: MN, 1SZ |
 |
Organism: Xenopus laevis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2003-02-18 Classification: STRUCTURAL PROTEIN/DNA Ligands: MN, IMT, PYB, ABU, BAL, DIB |
 |
Organism: Xenopus laevis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2003-02-18 Classification: STRUCTURAL PROTEIN/DNA Ligands: MN, IMT, PYB, ABU, BAL, DIB |
 |
Nmr Structure Of A 1:1 Complex Of Polyamide (Im-Py-Beta-Im-Beta-Im-Py-Beta-Dp) With The Tridecamer Dna Duplex 5'-Ccaaagagaagcg-3'
|

