Search Count: 15
 |
Human Mature Large Subunit Of The Ribosome With Eif6 And Homoharringtonine Bound
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:1.67 Å Release Date: 2023-03-08 Classification: TRANSLATION Ligands: MG, K, NA, HMT, SPM, EPE, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: RIBOSOME Ligands: K, MG, ZN |
 |
Cryo-Em Structure Of The Human Mtlsu Assembly Intermediate Upon Mrm2 Depletion - Class 4
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: RIBOSOME Ligands: MG, ZN |
 |
Cryo-Em Structure Of The Human Mtlsu Assembly Intermediate Upon Mrm2 Depletion - Class 1
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: RIBOSOME Ligands: ZN |
 |
Udp-Glucose Glycoprotein Glucosyltransferase From Chaetomium Thermophilum Soaked With K2Pti6
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:3.48 Å Release Date: 2017-07-26 Classification: TRANSFERASE Ligands: IOD, PT, CA |
 |
Udp-Glucose Glycoprotein Glucosyltransferase From Chaetomium Thermophilum (Open Conformation)
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:3.48 Å Release Date: 2017-07-26 Classification: TRANSFERASE Ligands: NAG, CA, EDO |
 |
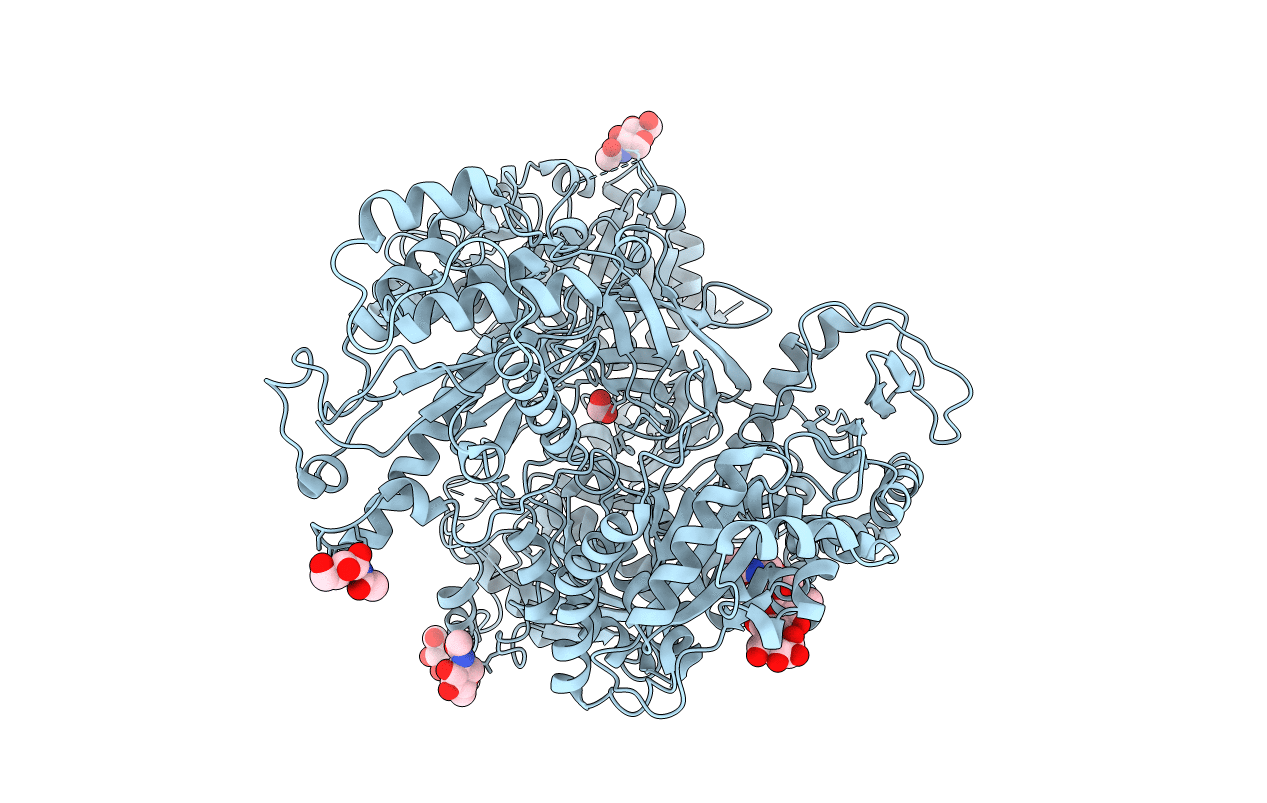
Udp-Glucose Glycoprotein Glucosyltransferase From Chaetomium Thermophilum (Closed Form)
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:4.40 Å Release Date: 2017-07-26 Classification: TRANSFERASE Ligands: NAG, CA, EDO |
 |
Udp-Glucose Glycoprotein Glucosyltransferase From Chaetomium Thermophilum Double Mutant D611C:G1050C
Organism: Chaetomium thermophilum var. thermophilum dsm 1495
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2017-07-26 Classification: TRANSFERASE Ligands: NAG, FMT |
 |
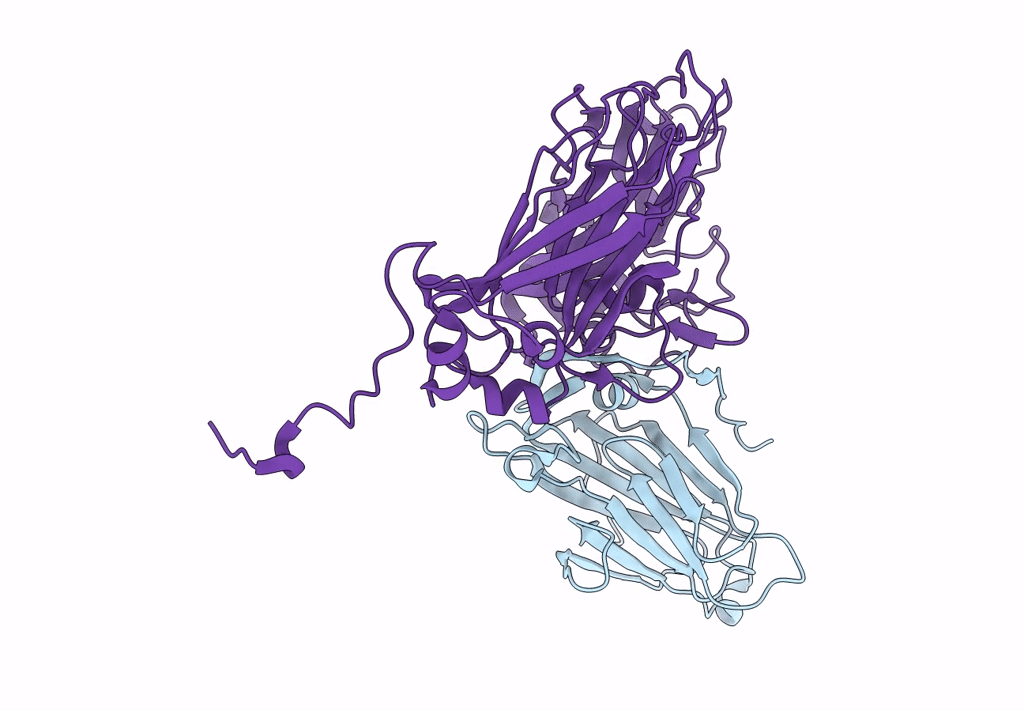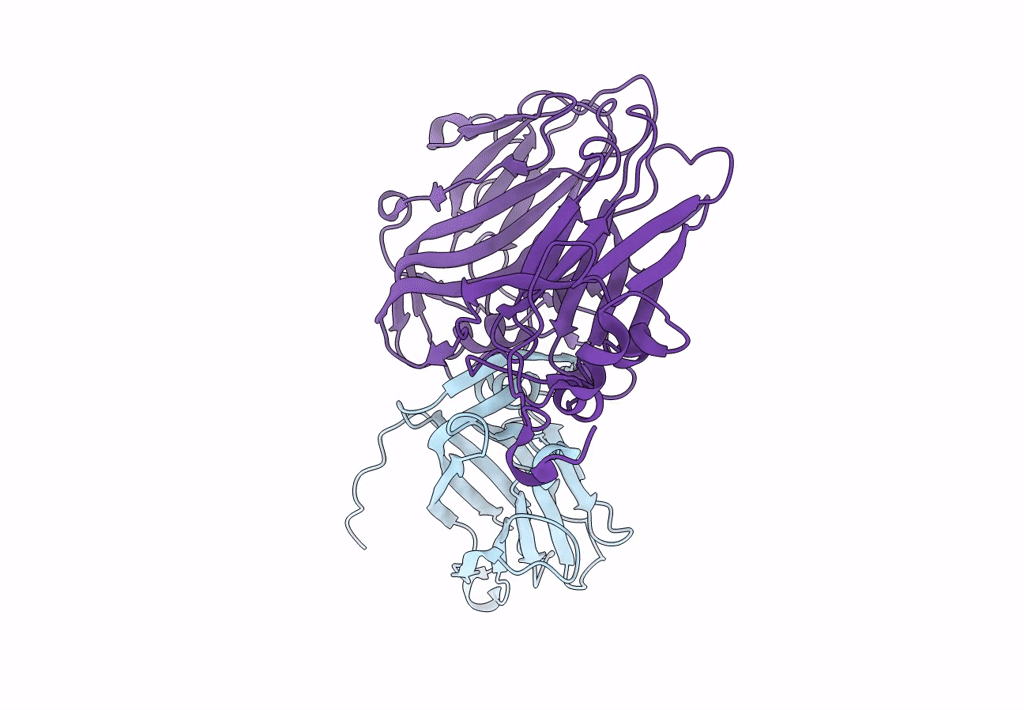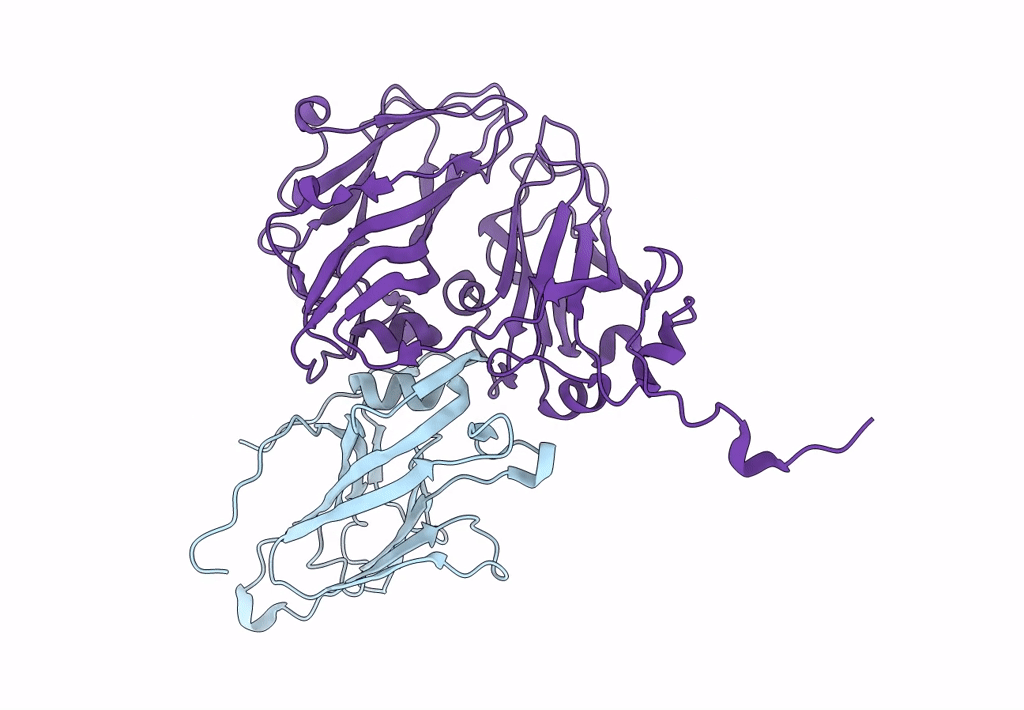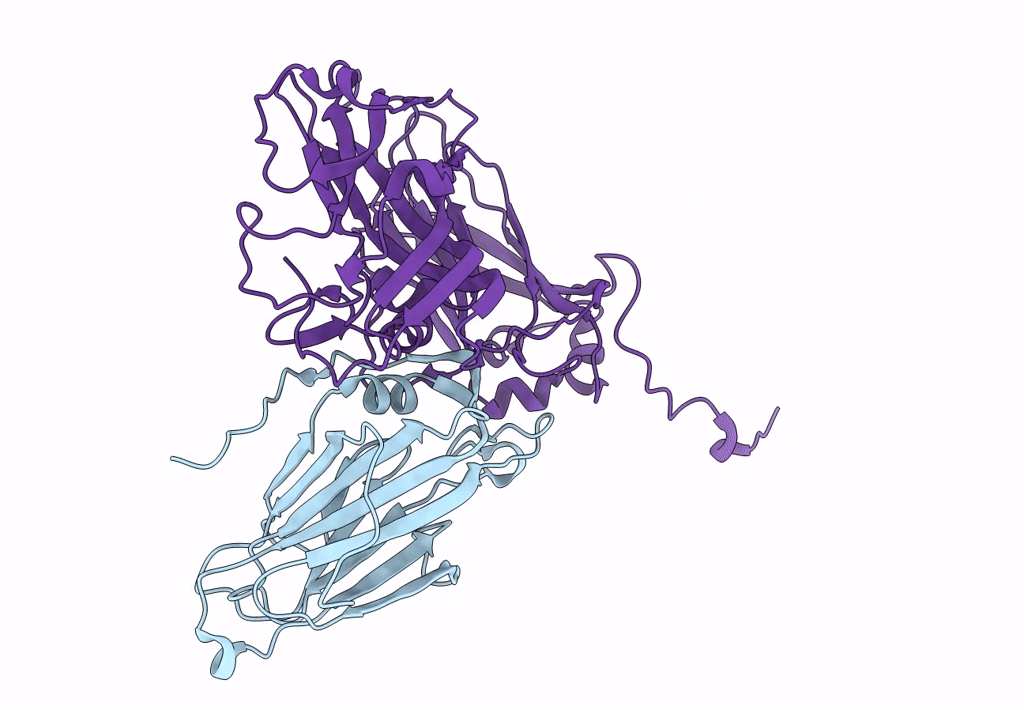
Pseudorabies Virus (Prv) Nuclear Egress Complex Proteins Fitted As A Hexameric Lattice Into A Sub-Tomogram Average Derived From Focused- Ion Beam Milled Lamellae Electron Cryo-Microscopic Data
Organism: Suid herpesvirus 1
Method: ELECTRON MICROSCOPY Resolution:35.00 Å Release Date: 2016-03-16 Classification: VIRAL PROTEIN Ligands: ZN, CL |
 |
Organism: Cowpea mosaic virus
Method: ELECTRON MICROSCOPY Resolution:3.44 Å Release Date: 2015-12-23 Classification: VIRUS |
 |
Electron Cryo-Microscopy Of Cowpea Mosaic Virus (Cpmv) Empty Virus Like Particle (Evlp)
Organism: Cowpea mosaic virus
Method: ELECTRON MICROSCOPY Resolution:3.04 Å Release Date: 2015-12-23 Classification: VIRUS |
 |
Organism: Suid herpesvirus 1
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-12-23 Classification: VIRAL PROTEIN Ligands: ZN, CL |
 |
Organism: Enterobacteria phage ms2
Method: ELECTRON MICROSCOPY Resolution:39.00 Å Release Date: 2013-07-17 Classification: VIRUS |
 |
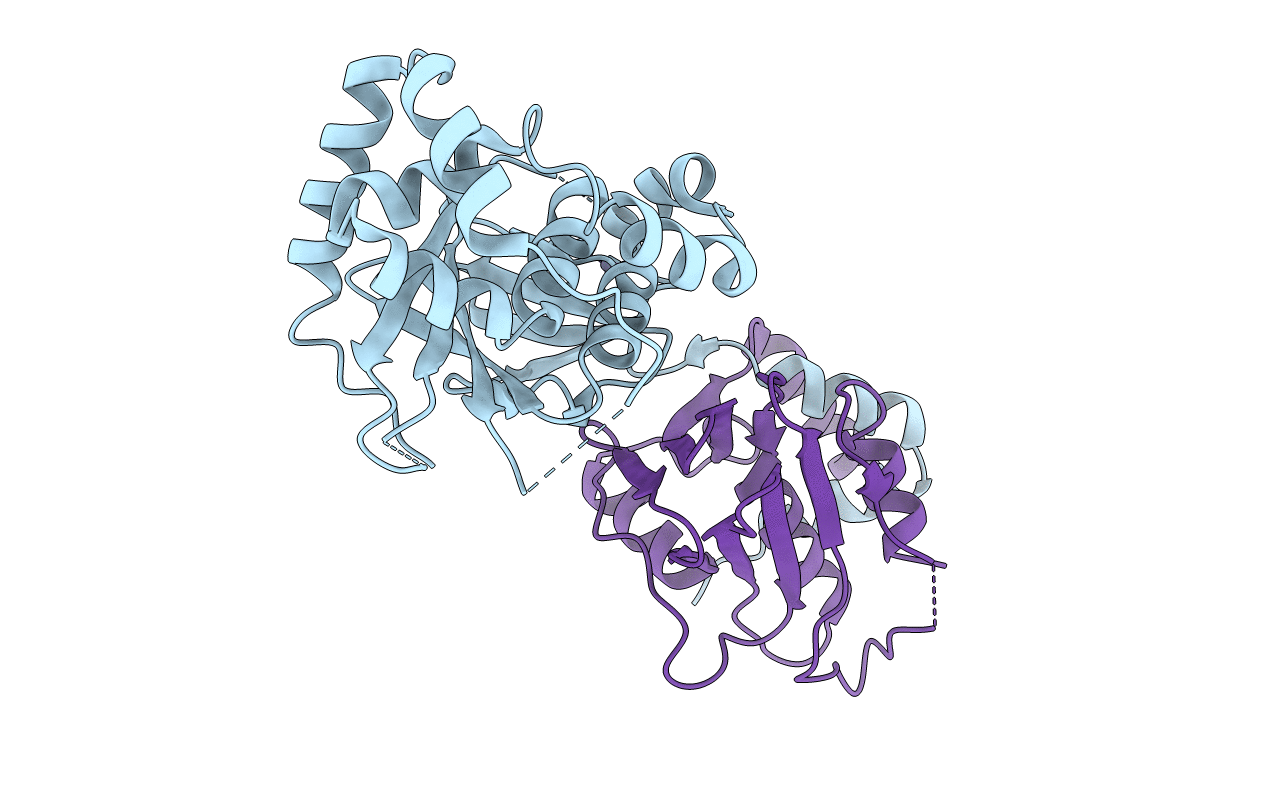
Organism: Schmallenberg virus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2013-05-01 Classification: VIRAL PROTEIN |
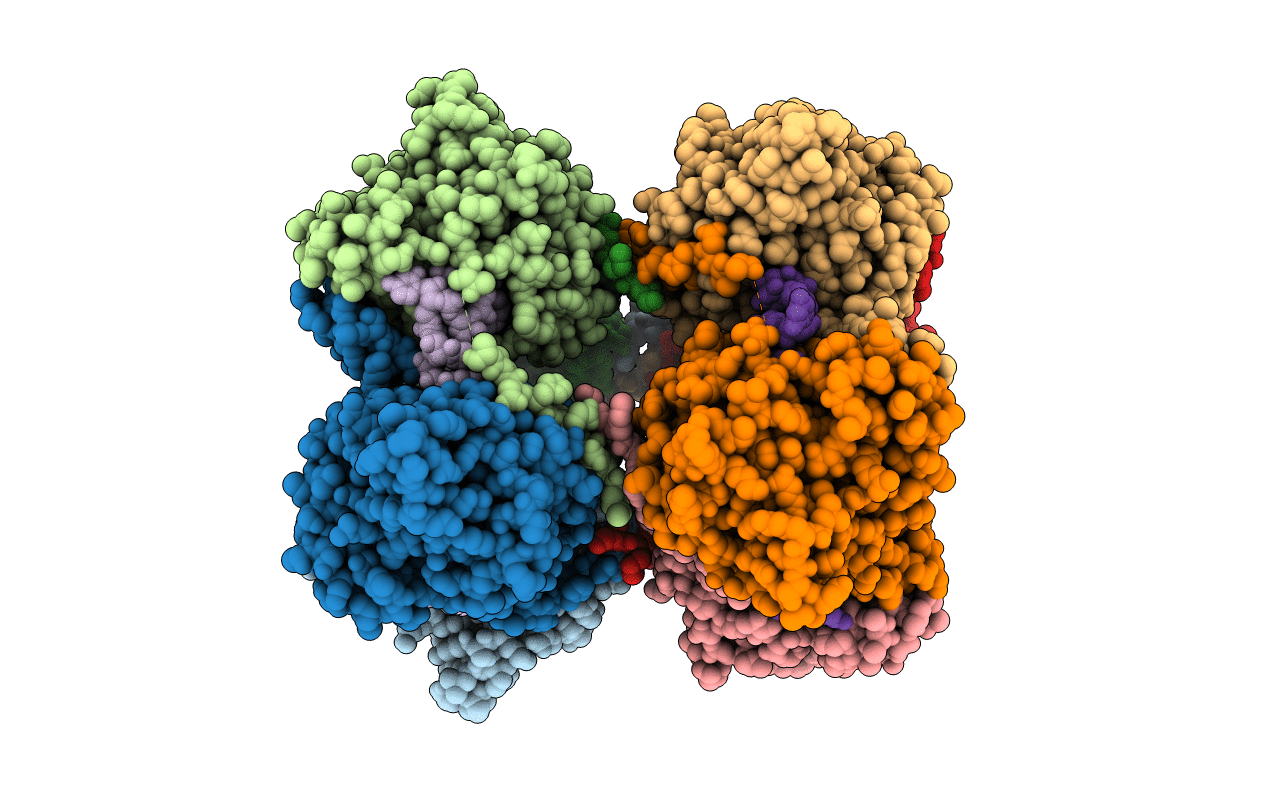 |
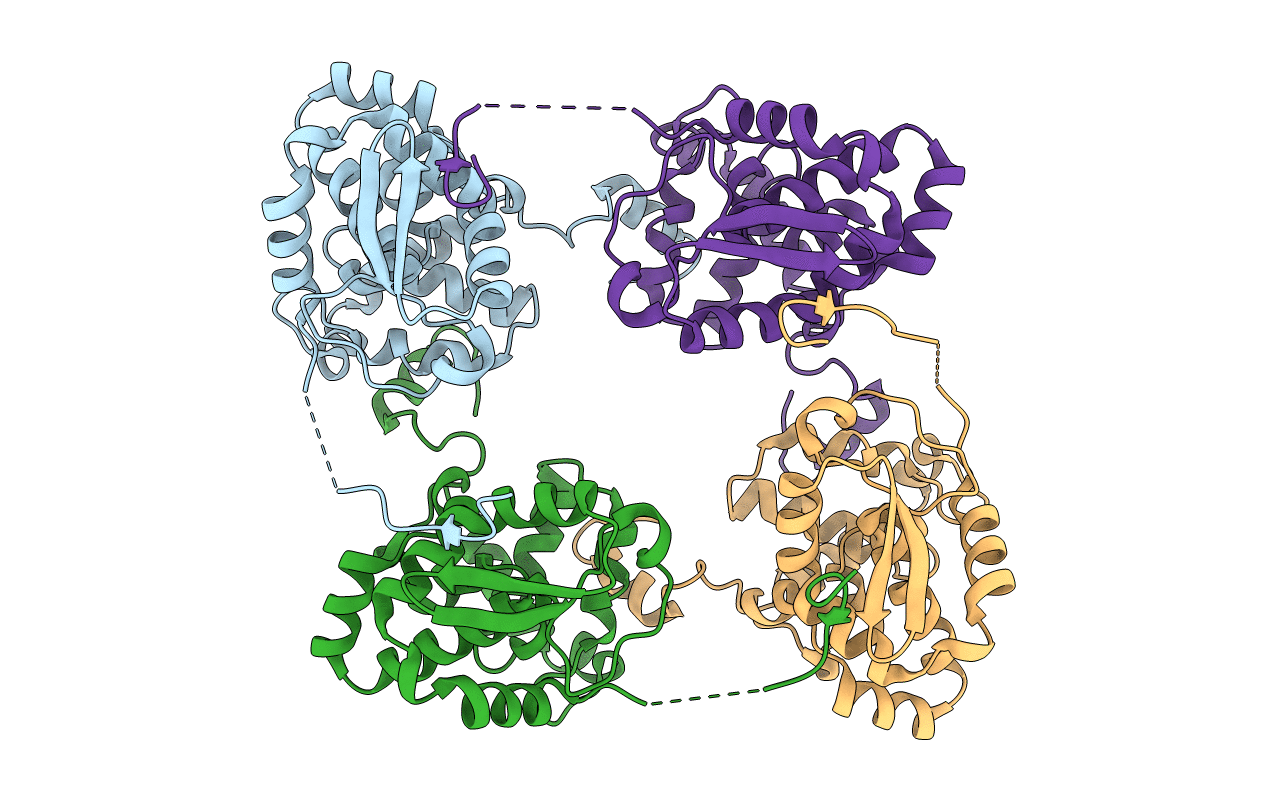
Crystal Structure Of The Nucleocapsid Protein From Bunyamwera Virus Bound To Rna
Organism: Bunyamwera virus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2013-05-01 Classification: VIRAL PROTEIN/RNA |

