Search Count: 355
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Actinosynnema pretiosum subsp. auranticum
Method: SOLUTION NMR Release Date: 2025-11-26 Classification: ANTIMICROBIAL PROTEIN |
 |
An Antibiotic Biosynthesis Monooxygenase Family Protein From Streptomyces Sp. Ma37
Organism: Streptomyces sp. ma37
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: ANTIBIOTIC |
 |
Crystal Structure Of A Polyketide Abm/Scha-Like Domain-Containing Protein Whie-Orfi From Streptomyces Coelicolor
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: ANTIBIOTIC |
 |
Organism: Henipavirus nipahense, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
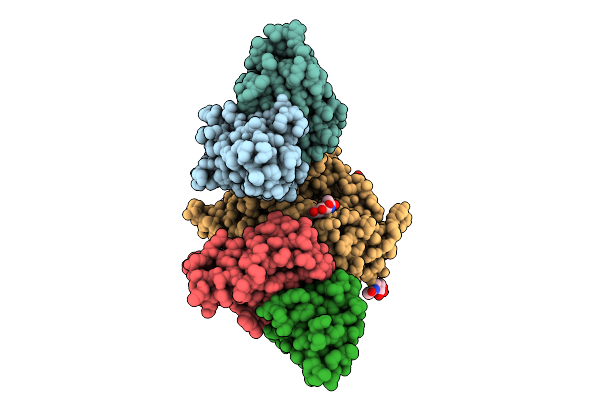 |
Organism: Henipavirus nipahense, Mesocricetus auratus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
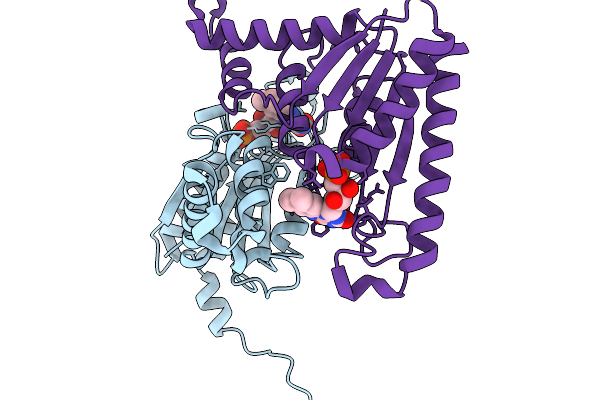 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: FLAVOPROTEIN Ligands: FMN |
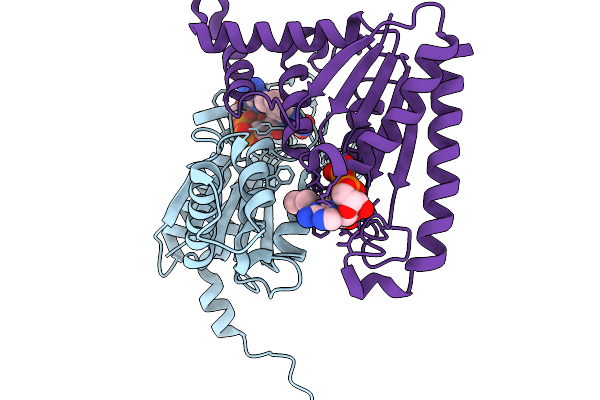 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: FLAVOPROTEIN Ligands: FAD |
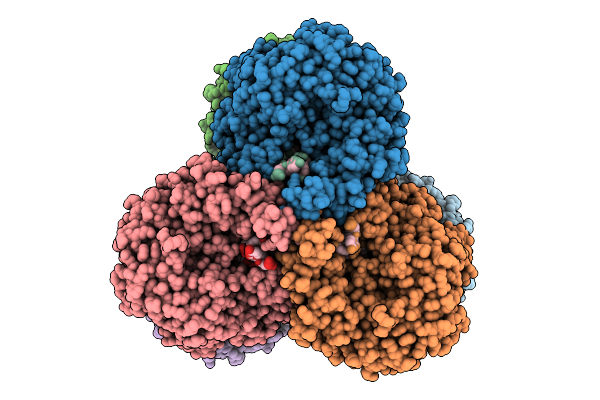 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN/DNA Ligands: DCP, MN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Sfts virus hb29, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: NUCLEAR PROTEIN/RNA |
 |
Organism: Streptomyces armeniacus
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: FE2, ILE |
 |
Organism: Streptomyces sp. z26
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: FE2, A1EAJ |
 |
Organism: Streptomyces sp. z26
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: FE2, A1EAI |
 |
Organism: Streptomyces sp. z26
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: FE2, A1EAJ |
 |
Organism: Streptomyces sp. z26
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: FE2, ILE |
 |
Organism: Streptomyces sp. z26
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: FE2, ILE |
 |
Organism: Streptomyces sp. nth33
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: FE2 |
 |
Organism: Streptomyces sp. nth33
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: FE2, H4B |

