Search Count: 28
 |
Organism: Entacmaea quadricolor
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: FLUORESCENT PROTEIN Ligands: EDO |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-19 Classification: TRANSCRIPTION Ligands: GOL, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: MG, ANP, IXC |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: MG, ANP, IXR |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: IMMUNE SYSTEM Ligands: NAG, Y01 |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: IMMUNE SYSTEM Ligands: Y01 |
 |
Structure Of The Human Ige-Fc Bound To Its High Affinity Receptor Fc(Epsilon)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: ANTIMICROBIAL PROTEIN Ligands: NAG |
 |
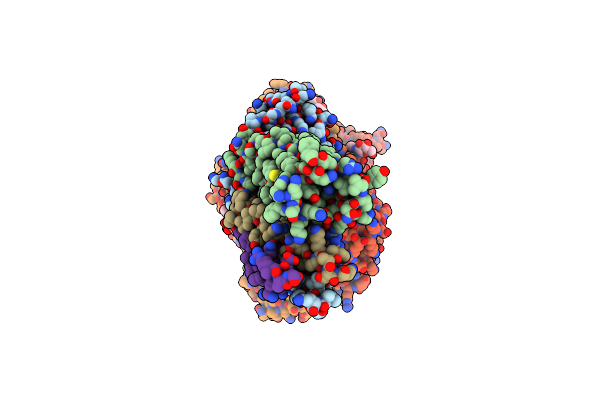
Structure Of The Ige-Fc Bound To Its High Affinity Receptor Fc(Epsilon)Ri State2
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: MEMBRANE PROTEIN Ligands: NAG, Y01 |
 |
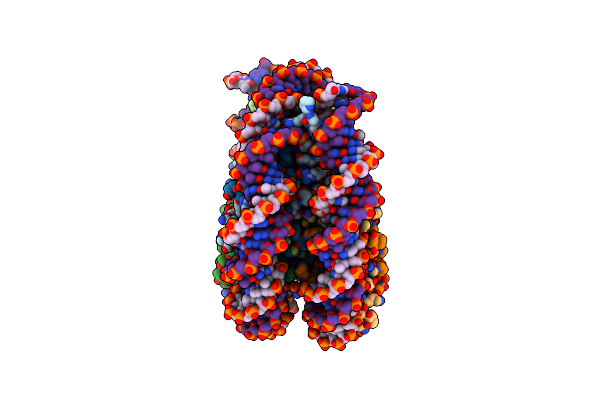
Structure Of The Ige-Fc Bound To Its High Affinity Receptor Fc(Epsilon)Ri State3
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: MEMBRANE PROTEIN Ligands: NAG, Y01 |
 |
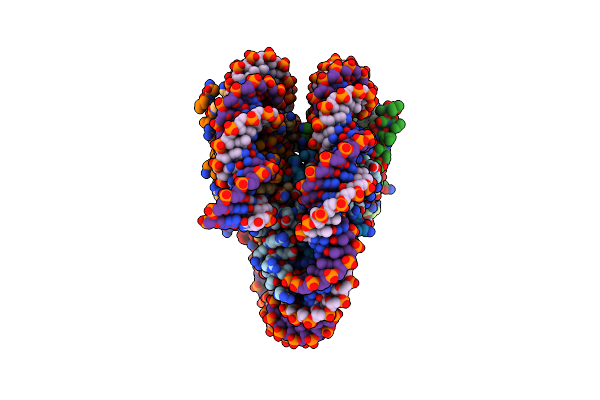
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: DNA BINDING PROTEIN/DNA |
 |
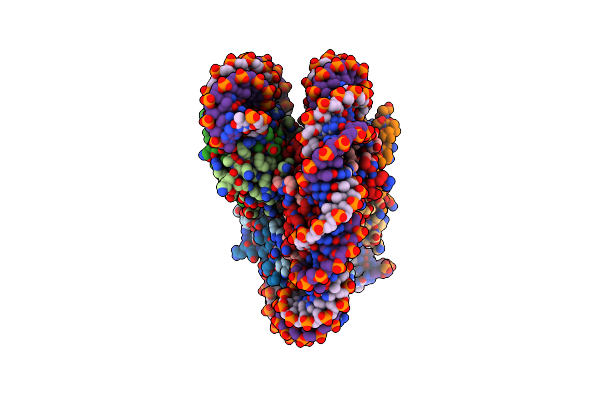
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Entacmaea quadricolor
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-07-27 Classification: FLUORESCENT PROTEIN Ligands: EDO, SO4, ACT |
 |
Organism: Entacmaea quadricolor
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2022-07-27 Classification: FLUORESCENT PROTEIN |
 |
Organism: Entacmaea quadricolor
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-07-27 Classification: FLUORESCENT PROTEIN Ligands: EDO |
 |
Organism: Entacmaea quadricolor
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-07-27 Classification: FLUORESCENT PROTEIN Ligands: EDO |
 |
Organism: Streptomyces platensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-07-24 Classification: OXIDOREDUCTASE Ligands: ACT, MN |
 |
Organism: Streptomyces platensis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2019-07-24 Classification: OXIDOREDUCTASE Ligands: ACT, MN, MZD |
 |
Organism: Streptomyces platensis
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2019-07-24 Classification: OXIDOREDUCTASE Ligands: ACT, MN, MYM |

