Search Count: 92
 |
N(5)-Hydroxyornithine:Cis-Anhydromevalonyl Coenzyme A-N(5)-Transacylase Sidf N-Terminal Domain
Organism: Aspergillus fumigatus (strain atcc mya-4609 / cbs 101355 / fgsc a1100 / af293)
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-08-14 Classification: TRANSFERASE |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 150 Micromolar Mg-132.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ALD, CL, NA, DMS, EDO |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 75 Micromolar Mg-132.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 150 Micromolar X77.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 75 Micromolar X77.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: CL, X77, EDO, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 500 Micromolar X77 Enantiomer R.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Millimolar X77 Enantiomer R.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 500 Micromolar X77 Enantiomer S.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: DMS, EDO, CL, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Millimolar X77 Enantiomer S.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: 9M5, EDO, ACT, DMS, CL, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Mm Mg-132, From An "Old" Crystal.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ALD, EDO, PEG, NA, CL |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Mm X77, From An "Old" Crystal.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ACT, EDO, CL, X77, NA |
 |
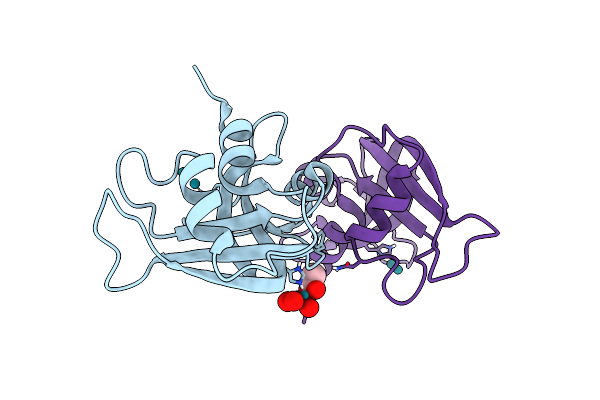
X-Ray Structure Of The Adduct Obtained Upon Reaction Of [Rh2(Ococh3)(Ococf3)3] With Rnase A (1)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-03-22 Classification: HYDROLASE Ligands: ACT, F3I, RH |
 |
X-Ray Structure Of The Adduct Obtained Upon Reaction Of [Rh2(Ococh3)(Ococf3)3] With Rnase A (2)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-03-22 Classification: HYDROLASE Ligands: F3I, RH |
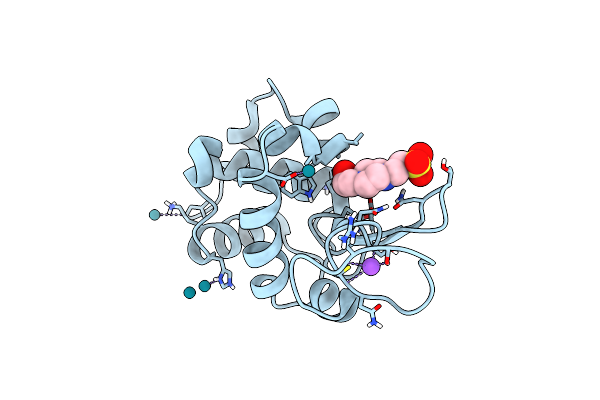 |
X-Ray Structure Of The Adduct Obtained Upon Reaction Of [Rh2(Ococh3)(Ococf3)3] With Hewl
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2023-03-22 Classification: HYDROLASE Ligands: EPE, NA, RH |
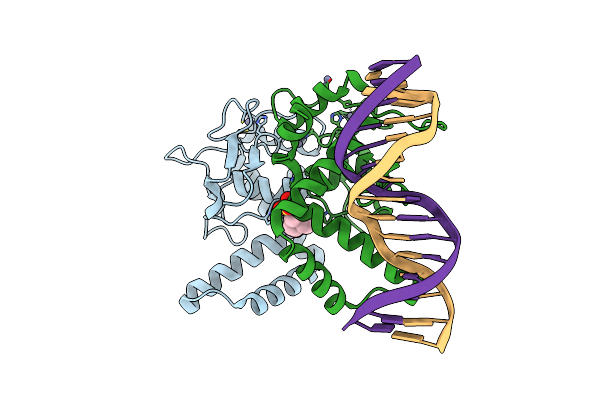 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-01-18 Classification: DNA BINDING PROTEIN Ligands: ZN, EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2023-01-18 Classification: DNA BINDING PROTEIN Ligands: ZN, EPE |
 |
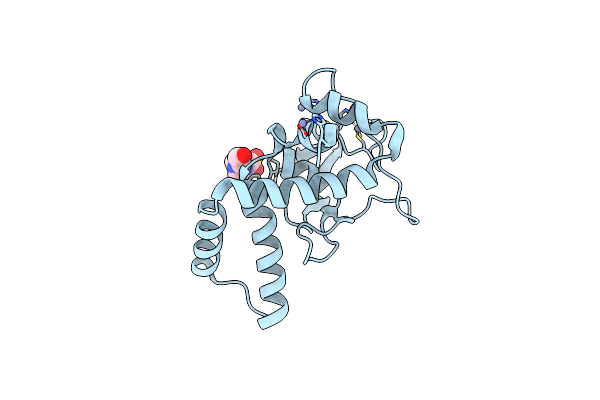
N(5)-Hydroxyornithine:Cis-Anhydromevalonyl Coenzyme A-N(5)-Transacylase Sidf
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2022-12-14 Classification: TRANSFERASE Ligands: GOL, SCN, K |
 |
X-Ray Structure Of The Adduct Obtained Upon Reaction Of [Cis-Rh2(Ococh3)2(Ococf3)2] With Rnase A (2)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2022-12-14 Classification: HYDROLASE Ligands: F3I, RHF |
 |
Orthorhombic Crystal Structure Of Ptg Cbm21 In Complex With Beta-Cyclodextrin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2022-11-02 Classification: SUGAR BINDING PROTEIN |
 |
Monoclinic Crystal Structure Of Ptg Cbm21 In Complex With Beta-Cyclodextrin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-11-02 Classification: SUGAR BINDING PROTEIN Ligands: GOL |

