Search Count: 101
 |
Crystal Structure Of Ctag From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: SO4, CO3 |
 |
Crystal Structure Of Ctag From Ruminiclostridium Cellulolyticum (P2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: GOL |
 |
Crystal Structure Of The Ctag_C11A Variant From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE |
 |
Crystal Structure Of The Ctag_C11A Variant From Ruminiclostridium Cellulolyticum (P2(1)2(1)2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: NI, IMD, TRS |
 |
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: SO4, CO3 |
 |
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum (P2(1)2(1)2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE |
 |
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum In Complex With Phba (P2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: HBA, PO4 |
 |
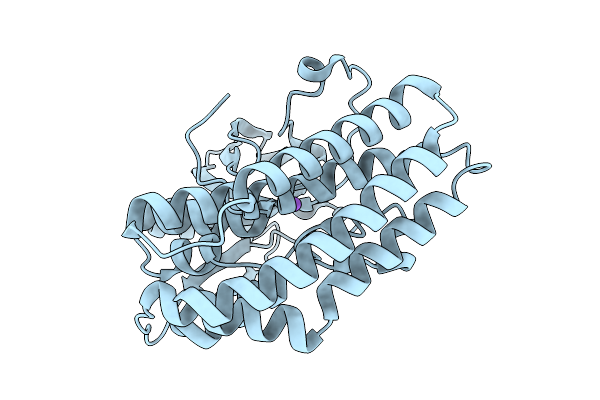
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum In Complex With Phba (P2(1)2(1)2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: HBA, GOL |
 |
Crystal Structure Of The Ctag_D144N Variant From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE |
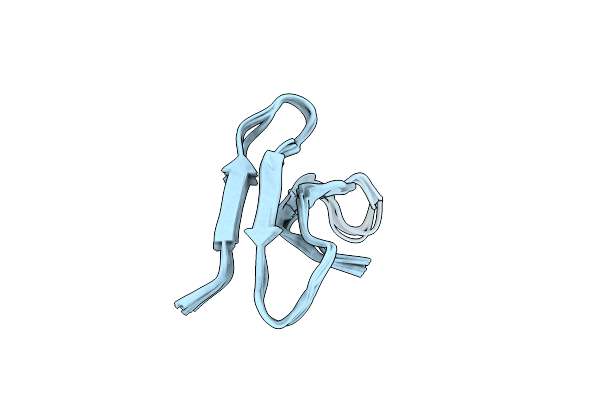 |
Solution Structure Of Sigma-S-Gviiia Conotoxin Extracted From Conus Geographus
Organism: Conus geographus
Method: SOLUTION NMR Release Date: 2025-05-07 Classification: NEUROPEPTIDE |
 |
Organism: Asterias rubens
Method: SOLUTION NMR Release Date: 2024-11-27 Classification: UNKNOWN FUNCTION |
 |
Organism: Asterias rubens
Method: SOLUTION NMR Release Date: 2024-11-27 Classification: UNKNOWN FUNCTION |
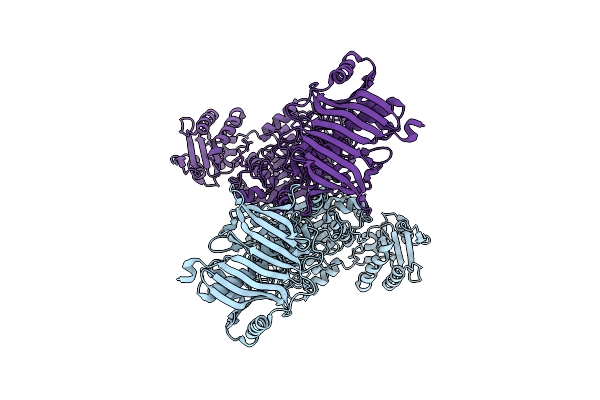 |
Organism: Mycetohabitans rhizoxinica
Method: ELECTRON MICROSCOPY Resolution:2.84 Å Release Date: 2024-04-03 Classification: TRANSFERASE |
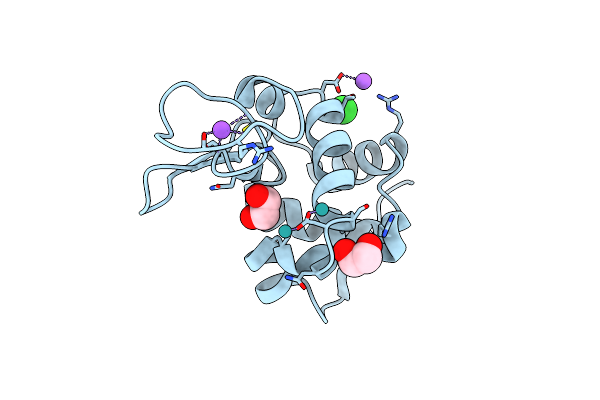 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2024-02-07 Classification: HYDROLASE Ligands: EDO, CL, NA, RU |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2024-01-24 Classification: HYDROLASE Ligands: EDO, ACT, RU, NA, CL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2024-01-24 Classification: HYDROLASE Ligands: EDO, RU, CL, NA |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-01-24 Classification: HYDROLASE Ligands: EDO, NA, CL, RU |
 |
Structure Of N-Terminal Of Schistosoma Japonicum Asparaginyl-Trna Synthetase
Organism: Schistosoma japonicum
Method: SOLUTION NMR Release Date: 2023-09-06 Classification: LIGASE |
 |
Organism: Tequatrovirus t4, Tequatrovirus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:4.98 Å Release Date: 2023-06-28 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Tequatrovirus, Tequatrovirus t4, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.53 Å Release Date: 2023-06-28 Classification: DNA BINDING PROTEIN |

