Search Count: 41
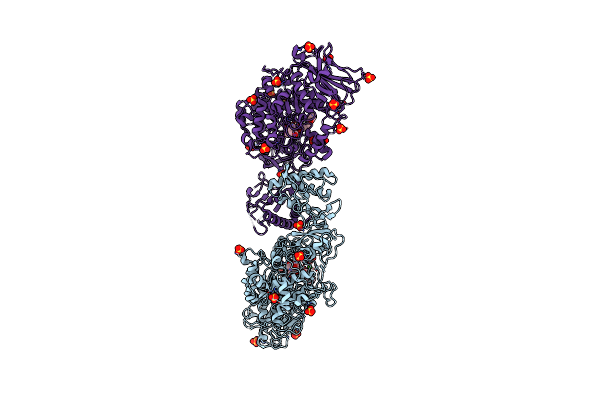 |
Catalytic Domain Of Gtfb In Complex With Inhibitor 2-[(2,4,5-Trihydroxyphenyl)Methylidene]-1-Benzofuran-3-One
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-06-14 Classification: TRANSFERASE/INHIBITOR Ligands: CA, XV5, BTB, SO4 |
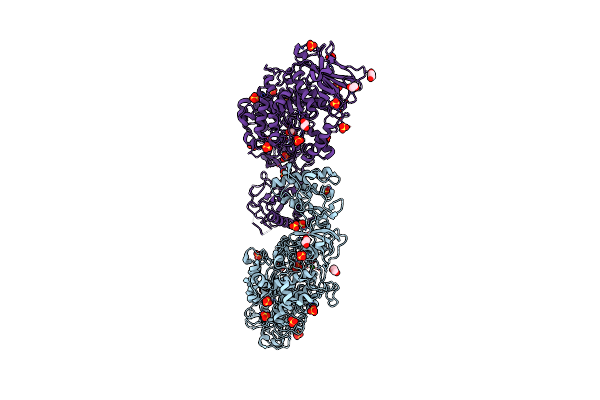 |
Structure Of The Catalytic Domain Of Streptococcus Mutans Gtfb In Tetragonal Space Group P4322
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-05-17 Classification: TRANSFERASE Ligands: CA, EDO, SO4, CL |
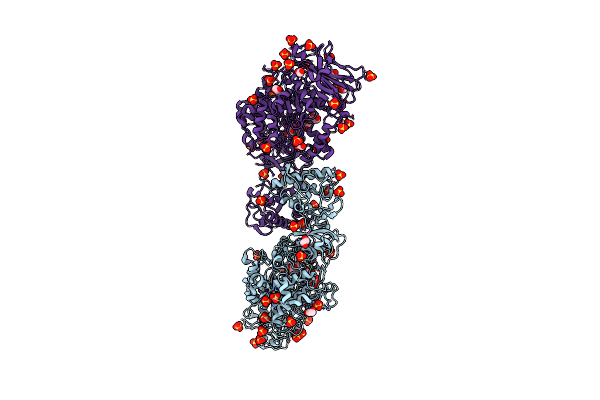 |
Structure Of The Catalytic Domain Of Streptococcus Mutans Gtfb Complexed To Acarbose In Tetragonal Space Group P4322
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-05-17 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: CA, EDO, SO4 |
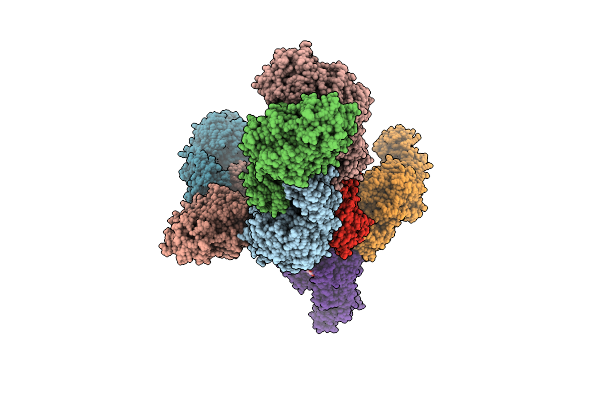 |
Structure Of The Catalytic Domain Of Streptococcus Mutans Gtfb Complexed To Acarbose In Orthorhombic Space Group P21212
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2023-05-17 Classification: TRANSFERASE Ligands: CA, SO4 |
 |
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2023-05-17 Classification: TRANSFERASE Ligands: MG, EDO |
 |
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-05-17 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-02 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2022-03-02 Classification: HYDROLASE Ligands: ATP, MG |
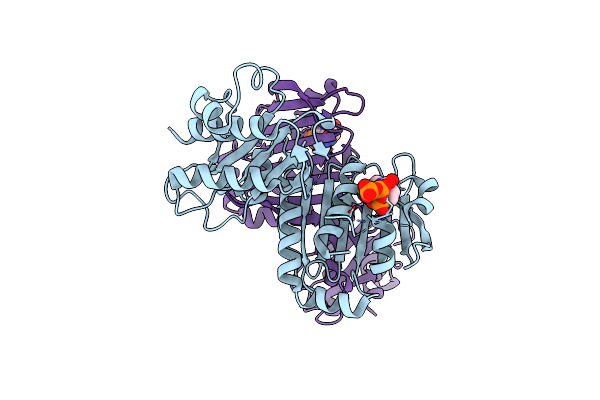 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-03-02 Classification: HYDROLASE Ligands: ATP, MG |
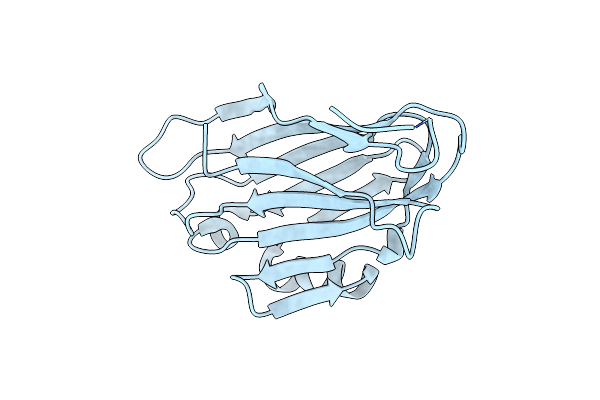 |
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-01-26 Classification: CELL ADHESION |
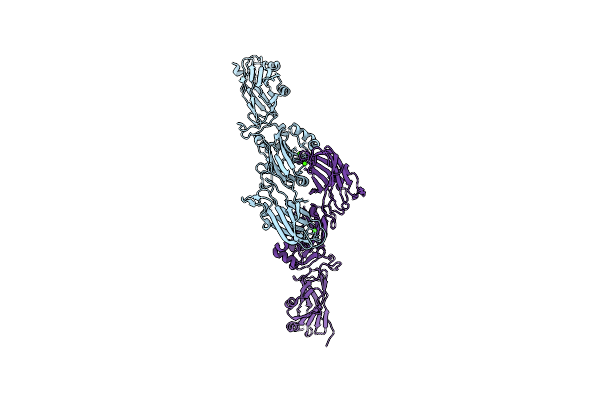 |
Organism: Streptococcus gordonii
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-09-08 Classification: CELL ADHESION Ligands: CA |
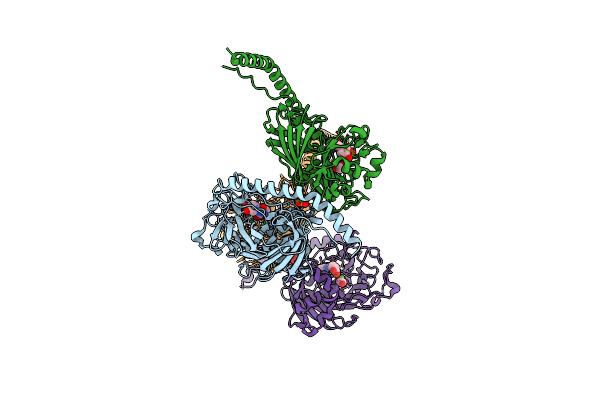 |
The Structure Of The Streptococcus Gordonii Surface Protein Sspb In Complex With Tev Peptide Provides Clues To The Adherence Of Oral Streptococcal Adherence To Salivary Agglutinin
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-09-16 Classification: CELL ADHESION Ligands: CA, SNZ, NA, SO4, EDO |
 |
The Structure Of The Streptococcus Gordonii Surface Protein Sspb In Complex With Tev Peptide Provides Clues To The Adherence Of Oral Streptococcal Adherence To Salivary Agglutinin
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-08-19 Classification: CELL ADHESION Ligands: SO4 |
 |
The Structure Of The Streptococcus Gordonii Surface Protein Sspb In Complex With Tev Peptide Provides Clues To The Adherence Of Oral Streptococcal Adherence To Salivary Agglutinin
Organism: Streptococcus gordonii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-08-12 Classification: CELL ADHESION Ligands: CA |
 |
The Structure Of The Streptococcus Gordonii Surface Protein Sspb In Complex With Tev Peptide Provides Clues To The Adherence Of Oral Streptococcal Adherence To Salivary Agglutinin
Organism: Streptococcus gordonii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-08-12 Classification: CELL ADHESION Ligands: MG, GOL |
 |
Crystal Structure Of A Tylonycteris Bat Coronavirus Hku4 Macrodomain In Complex With Adenosine Diphosphate Ribose (Adp-Ribose)
Organism: Bat coronavirus hku4
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2019-09-11 Classification: HYDROLASE Ligands: APR |
 |
Crystal Structure Of Tylonycteris Bat Coronavirus Hku4 Macrodomain In Complex With Nicotinamide Adenine Dinucleotide (Nad+)
Organism: Bat coronavirus hku4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-09-11 Classification: HYDROLASE Ligands: NAD |
 |
Crystal Structure Of A Tylonycteris Bat Coronavirus Hku4 Macrodomain In Complex With Adenosine Diphosphate Glucose (Adp-Glucose)
Organism: Bat coronavirus hku4
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-09-11 Classification: HYDROLASE Ligands: ADQ |
 |
The Structure Of The Variable Domain Of Streptococcus Intermedius Antigen I/Ii (Pas)
Organism: Streptococcus intermedius
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-07-17 Classification: CELL ADHESION Ligands: MG |
 |
The Structure Of The C-Terminal Domains (C123) Of Streptococcus Intermedius Antigen I/Ii (Pas)
Organism: Streptococcus intermedius
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-07-17 Classification: CELL ADHESION Ligands: CA |

