Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of The Endo-1,4-Glucanase, Rbcel1, In Complex With Cellobiose
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2014-12-31 Classification: HYDROLASE Ligands: TRS |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2013-04-03 Classification: HYDROLASE Ligands: TRS |
 |
Crystal Structure Of The Mono-Zinc Metallo-Beta-Lactamase Vim-4 From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2010-06-23 Classification: HYDROLASE Ligands: ZN, FLC, CIT |
 |
Organism: Aeromonas hydrophila
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2010-06-23 Classification: HYDROLASE Ligands: ZN, IFS, SO4, GOL |
 |
Organism: Aeromonas hydrophila
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2010-06-23 Classification: HYDROLASE Ligands: ZN, GOL, SO4, SDF |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2009-11-10 Classification: HYDROLASE/ANTIBIOTIC Ligands: PNM, GOL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-10-28 Classification: HYDROLASE Ligands: SO4, GOL, EDO |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2007-07-03 Classification: HYDROLASE Ligands: SO4, CO |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-07-03 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-07-03 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2007-07-03 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Citrobacter freundii
Method: SOLUTION NMR Release Date: 2003-02-18 Classification: HYDROLASE Ligands: ZN |
 |
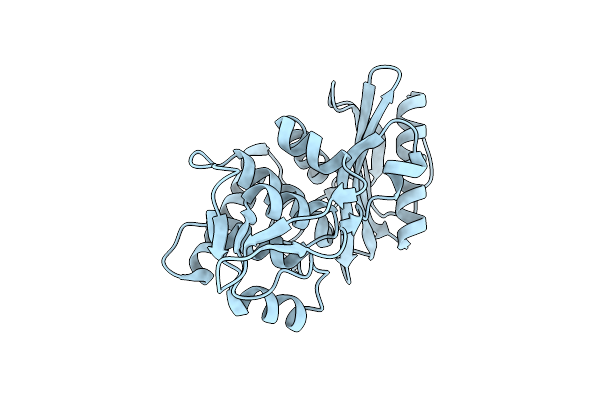
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2001-06-13 Classification: HYDROLASE |
 |
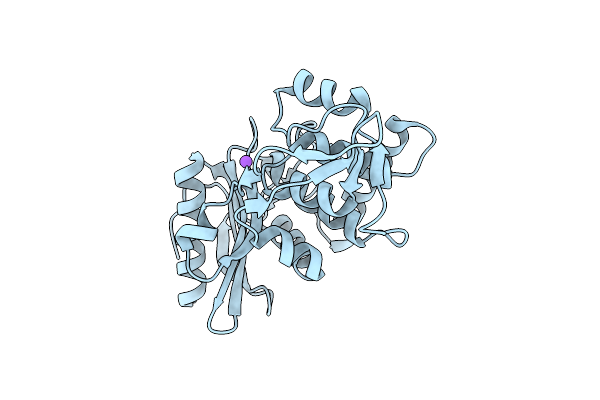
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2000-05-03 Classification: HYDROLASE |
 |
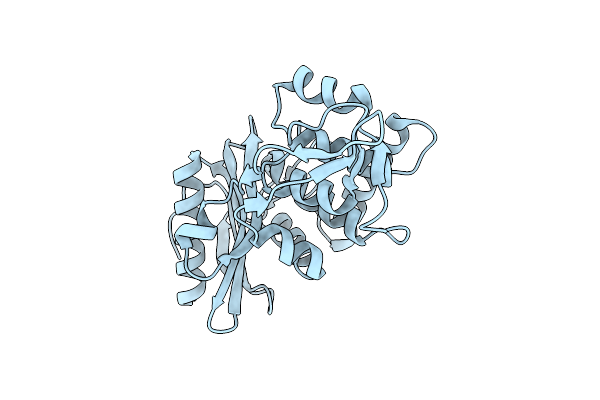
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2000-05-03 Classification: HYDROLASE |
 |
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2000-05-03 Classification: HYDROLASE |