Search Count: 48
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-10-18 Classification: FLUORESCENT PROTEIN |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2023-10-18 Classification: FLUORESCENT PROTEIN |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-09-27 Classification: FLUORESCENT PROTEIN |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2023-07-05 Classification: FLUORESCENT PROTEIN Ligands: GOL, SO4 |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2023-07-05 Classification: FLUORESCENT PROTEIN Ligands: GOL, SO4 |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2023-07-05 Classification: FLUORESCENT PROTEIN Ligands: GOL, SO4 |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2022-11-09 Classification: FLUORESCENT PROTEIN Ligands: SO4 |
 |
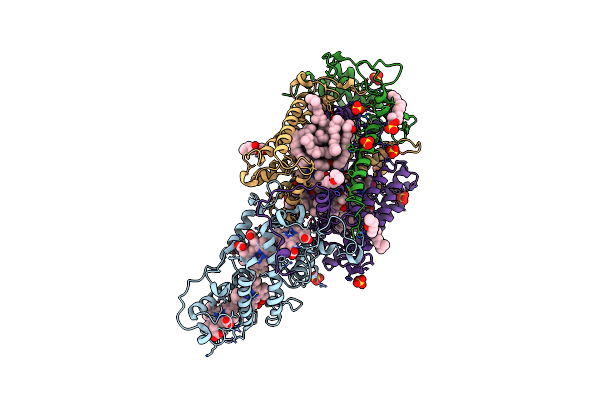
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 1 Ps Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
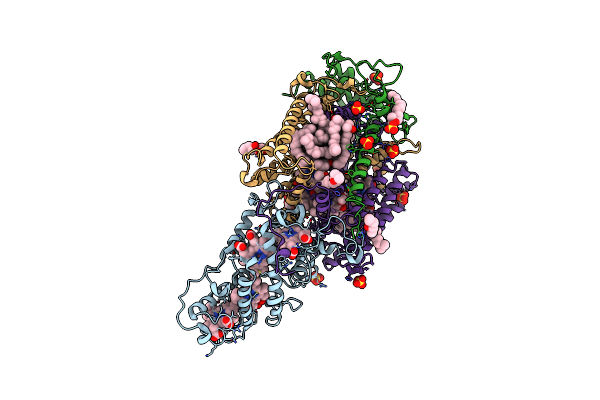
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 5 Ps (A) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
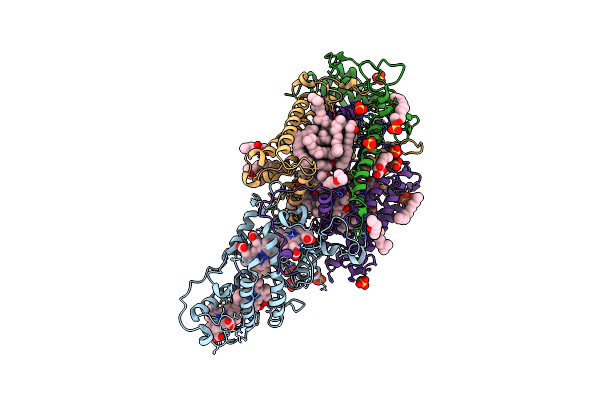
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 300 Ps (A) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
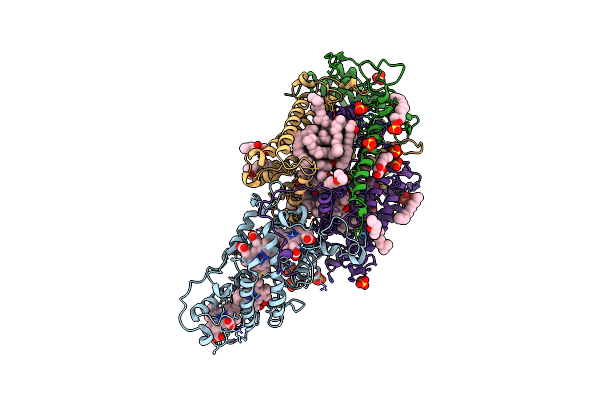
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 20 Ps Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
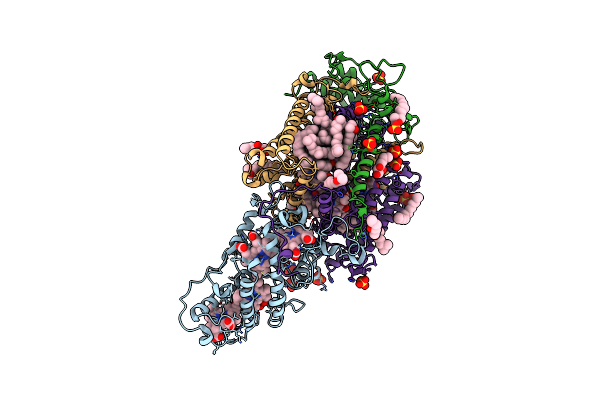
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 300 Ps (B) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 8 Us Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 5 Ps (B) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
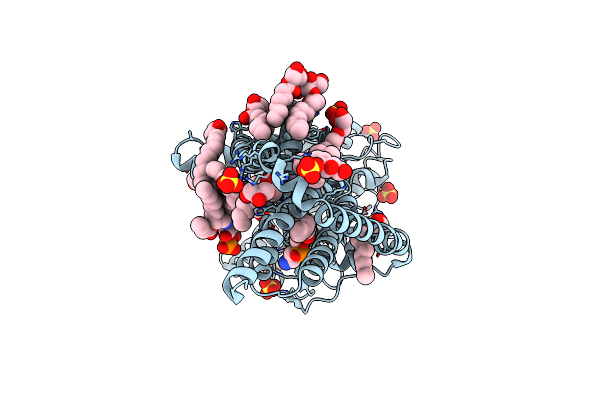 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-08-26 Classification: LIPID BINDING PROTEIN Ligands: SO4, OLB, PG4, 3PE |
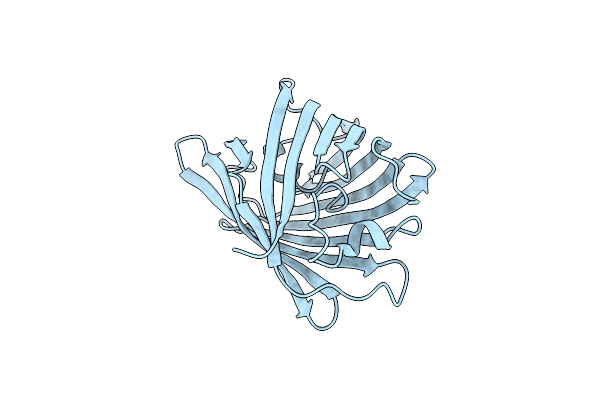 |
Xfel Structure Of The On State Of A Reversibly Photoswitching Fluorescent Protein Determined Using The Grease Injection Method
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2017-12-06 Classification: FLUORESCENT PROTEIN |
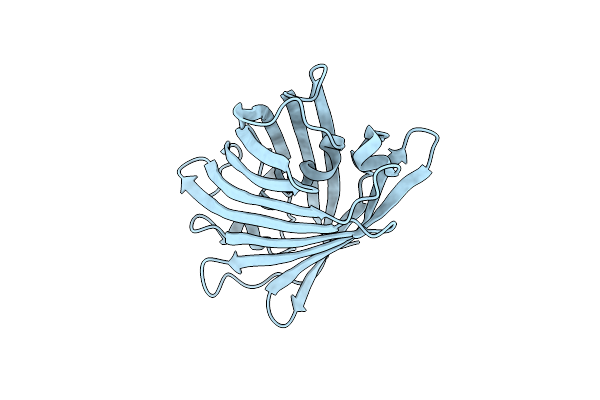 |
Xfel Structure Of The Off State Of A Reversibly Photoswitching Fluorescent Protein Determined Using The Droplet On Demand Injection Method
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2017-12-06 Classification: FLUORESCENT PROTEIN |
 |
Xfel Structure Of The Off State Of A Reversibly Photoswitching Fluorescent Protein Determined Using The Gdvn Injection Method
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2017-12-06 Classification: FLUORESCENT PROTEIN |
 |
Xfel Structure Of The On State Of A Reversibly Photoswitching Fluorescent Protein Determined Using The Droplet Injection Method
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2017-12-06 Classification: FLUORESCENT PROTEIN |
 |
From Macrocrystals To Microcrystals: A Strategy For Membrane Protein Serial Crystallography
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-08-16 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE2, MQ7, NS5 |

