Search Count: 81
 |
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (3 Picosecond Pump-Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (300 Ps Pump-Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (1 Nanosecond Pump-Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (3 Nanosecond Pump-Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-11-22 Classification: LYASE Ligands: FDA, SO4 |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (10 Nanosecond Pump-Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2023-11-22 Classification: LYASE Ligands: FDA, SO4 |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (30 Nanosecond Timepoint)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (1 Microsecond Pump-Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (10 Microsecond Pump Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (30 Microsecond Pump-Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (100 Microsecond Timpeoint)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Organism: Discosoma sp.
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-04-05 Classification: FLUORESCENT PROTEIN Ligands: PEG, MES, EDO, GOL |
 |
Organism: Discosoma sp.
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-04-05 Classification: FLUORESCENT PROTEIN Ligands: GOL, MES, PEG, EDO |
 |
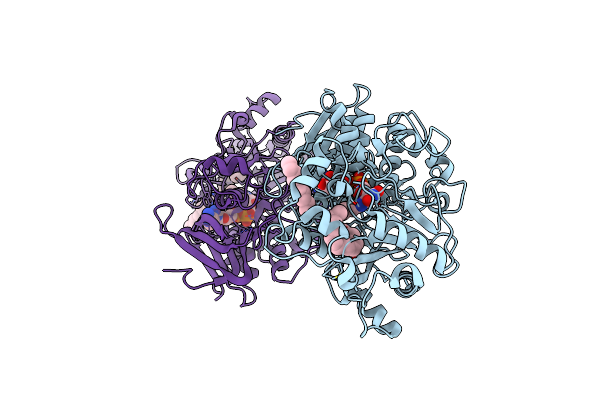
Difference-Refined Structure Of Fatty Acid Photodecarboxylase 20 Ps Following 400-Nm Laser Irradiation Of The Dark-State Determined By Sfx
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-14 Classification: OXIDOREDUCTASE Ligands: FAD, STE |
 |
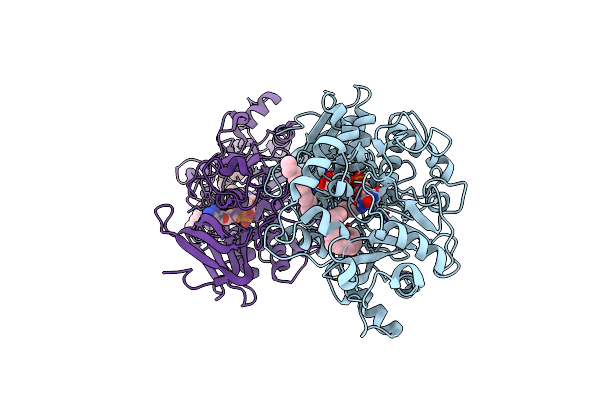
Difference-Refined Structure Of Fatty Acid Photodecarboxylase 900 Ps Following 400-Nm Laser Irradiation Of The Dark-State Determined By Sfx
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-14 Classification: OXIDOREDUCTASE Ligands: FAD, STE |
 |
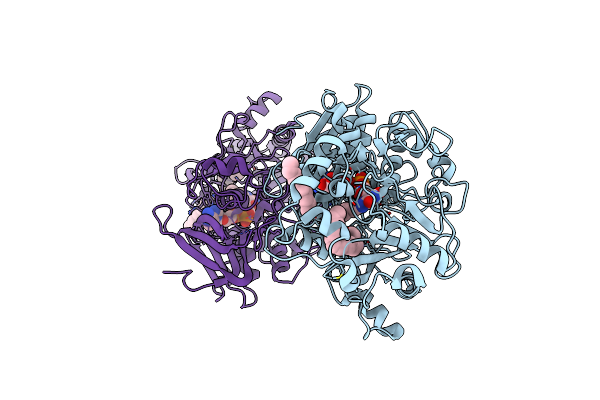
Difference-Refined Structure Of Fatty Acid Photodecarboxylase 300 Ns Following 400-Nm Laser Irradiation Of The Dark-State Determined By Sfx
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-14 Classification: OXIDOREDUCTASE Ligands: FAD, STE |
 |
Difference-Refined Structure Of Fatty Acid Photodecarboxylase 2 Microsecond Following 400-Nm Laser Irradiation Of The Dark-State Determined By Sfx
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-09-14 Classification: OXIDOREDUCTASE Ligands: FAD, STE |
 |
Mosquitocidal Cry11Aa Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Aa-Y449F Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Aa-E583Q Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2022-07-27 Classification: TOXIN |

