Search Count: 73
 |
2.03 Angstrom Resolution Crystal Structure Of As-Isolated Katg From Mycobacterium Tuberculosis With An Myw-Ooh Cofactor
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2024-10-23 Classification: OXIDOREDUCTASE Ligands: HEM, NA, GOL, ACT |
 |
2.35-Angstrom Resolution Intermediate Crystal Structure Of Katg From Mycobacterium Tuberculosis With An Myw-Ooh Cofactor Soaked With Peroxide For 5 Minutes
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-10-23 Classification: OXIDOREDUCTASE Ligands: HEM, OXY, NA, ACT, GOL |
 |
2.30 Angstrom Resolution Intermediate Crystal Structure Of Katg From Mycobacterium Tuberculosis With An Myw-Ooh Cofactor Soaked With Peroxide For 1 Minute
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-10-23 Classification: OXIDOREDUCTASE Ligands: HEM, NA, GOL |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2024-10-16 Classification: LYASE Ligands: ZN, MLA |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-10-16 Classification: LYASE Ligands: ZN |
 |
1.79 Angstrom Resolution Crystal Structure Of Katg From Mycobacterium Tuberculosis With An Myw Cofactor After Heat Incubation For 60 Minutes
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2024-09-11 Classification: OXIDOREDUCTASE |
 |
2.25 Angstrom Resolution Crystal Structure Of As-Isolated Katg From Mycobacterium Tuberculosis With An Myw Cofactor
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE Ligands: HEM, ACT |
 |
Time-Resolved Sfx-Xfel Crystal Structure Of Cyp121 Bound With Cyy Reacted With Peracetic Acid For 200 Milliseconds
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: SO4, HEM, YTT, PEO |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: SO4, HEM, YTT |
 |
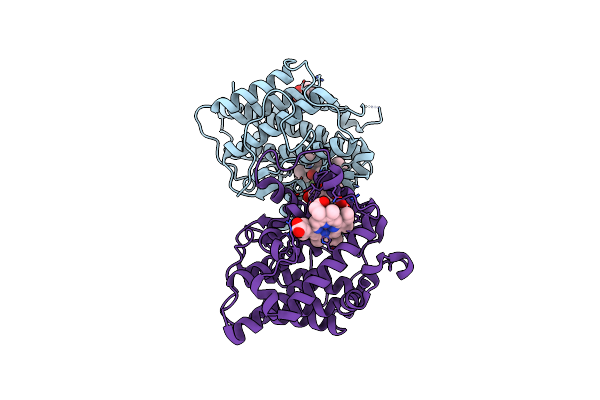
A 1.89-A Resolution Substrate-Bound Crystal Structure Of Heme-Dependent Tyrosine Hydroxylase From S. Sclerotialus
Organism: Streptomyces sclerotialus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2021-03-31 Classification: OXIDOREDUCTASE Ligands: HEM, TYR, BTB, TRS |
 |
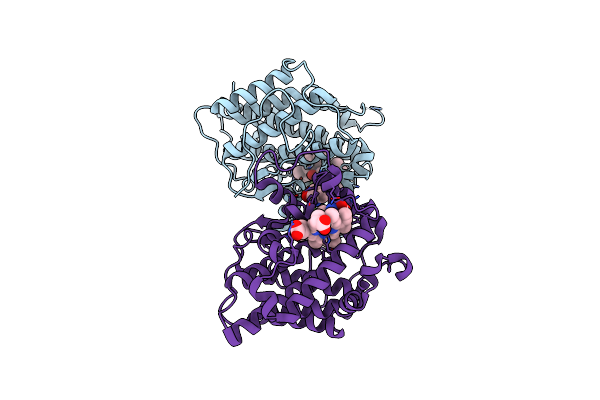
A 1.68-A Resolution 3-Fluoro-L-Tyrosine Bound Crystal Structure Of Heme-Dependent Tyrosine Hydroxylase
Organism: Streptomyces sclerotialus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2021-03-31 Classification: OXIDOREDUCTASE Ligands: HEM, TRS, BTB, YOF |
 |
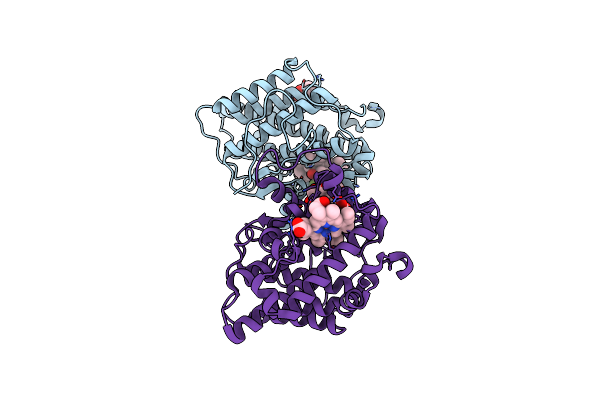
A 1.84-A Resolution Crystal Structure Of Heme-Dependent L-Tyrosine Hydroxylase In Complex With 3-Fluoro-L-Tyrosine And Cyanide
Organism: Streptomyces sclerotialus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-03-31 Classification: OXIDOREDUCTASE Ligands: HEM, YOF, CYN, TRS |
 |
A 1.58-A Resolution Crystal Structure Of Ferric-Hydroperoxo Intermediate Of L-Tyrosine Hydroxylase In Complex With 3-Fluoro-L-Tyrosine
Organism: Streptomyces sclerotialus
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-03-31 Classification: OXIDOREDUCTASE Ligands: HEM, TRS, YOF, PEO, BTB |
 |
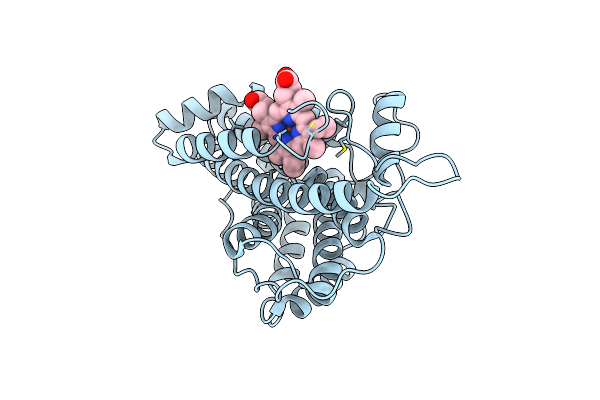
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-03-10 Classification: OXIDOREDUCTASE Ligands: HEC |
 |
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2021-03-10 Classification: OXIDOREDUCTASE Ligands: HEC |
 |
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2021-03-10 Classification: OXIDOREDUCTASE Ligands: HEC |
 |
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2021-03-10 Classification: OXIDOREDUCTASE Ligands: HEC |
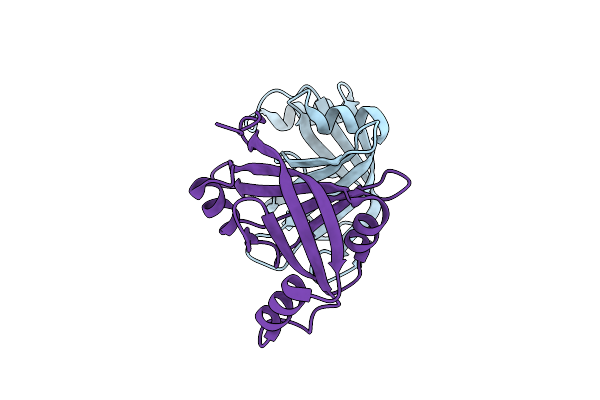 |
Organism: Streptococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-02-10 Classification: OXIDOREDUCTASE |
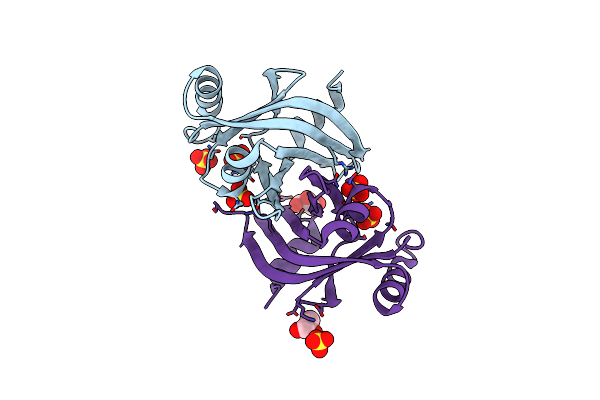 |
1.98 A Resolution Crystal Structure Of Group A Streptococcus H111A Hupz-V5-His6
Organism: Streptococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2021-02-10 Classification: OXIDOREDUCTASE Ligands: SO4, GOL |
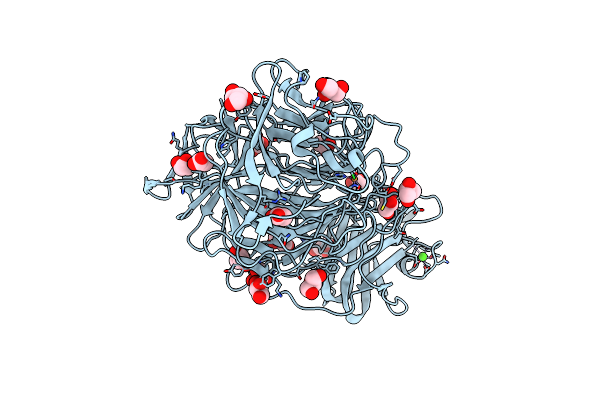 |
The 1.23 Angstrom Crystal Structure Of Galactose Oxidase Variant With Genetically Incorporated Cl2-Tyr272
Organism: Gibberella zeae
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2021-02-03 Classification: OXIDOREDUCTASE Ligands: CU, CA, ACT, GOL |

