Search Count: 107
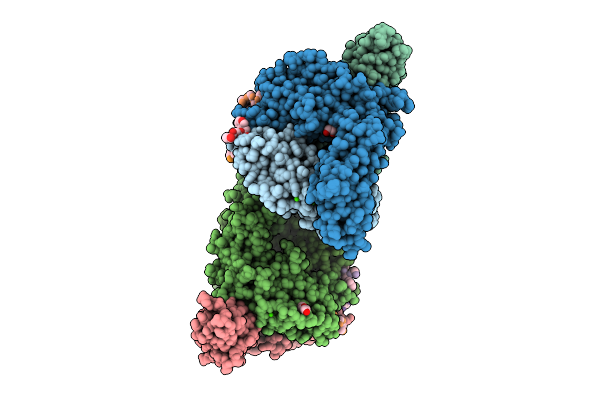 |
Vibrio Cholerae Protein Frha Peptid-Binding Domain And Adjacent Split Domain (S1127-F1439) In Complex With Peptide Agwtd X-Ray Crystallography Structure
Organism: Vibrio cholerae o395, Vibrio cholerae
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: CA, PEG, EDO |
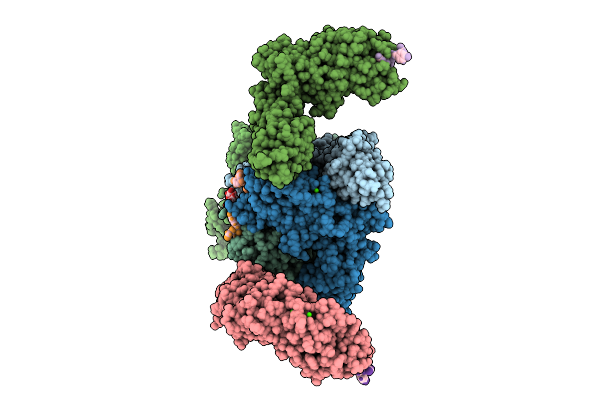 |
Vibrio Cholerae Protein Frha Peptid-Binding Domain And Adjacent Split Domain (S1127-F1439) In Complex With Peptide Agytd X-Ray Crystallography Structure
Organism: Vibrio cholerae o395, Vibrio cholerae
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: CA |
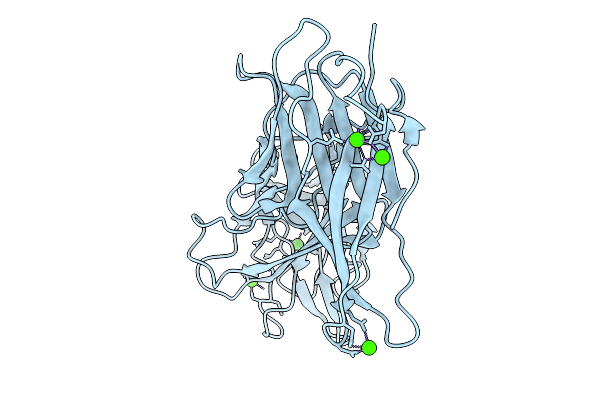 |
Crystal Structure Of Aeromonas Salmonicida Putative Carbohydrate Binding Module And Split Domain
Organism: Aeromonas salmonicida subsp. salmonicida a449
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: CELL ADHESION Ligands: CA |
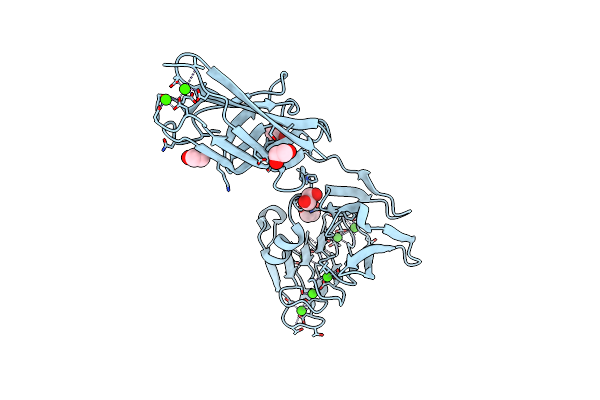 |
Crystal Structure Of Repeats-In-Toxin-Like Domain From Aeromonas Hydrophila
Organism: Aeromonas hydrophila
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-05-14 Classification: CELL ADHESION Ligands: EDO, CA |
 |
Crystal Structure Of Vwfa Domain From Large Adhesion Protein Of Aeromonas Hydrophila
Organism: Aeromonas hydrophila
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-05-14 Classification: CELL ADHESION Ligands: CA, GOL |
 |
Organism: Marinomonas primoryensis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-09-01 Classification: PROTEIN BINDING Ligands: 1PE, CA |
 |
Organism: Marinomonas primoryensis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-09-01 Classification: PROTEIN BINDING Ligands: CA, NA, 1PE |
 |
Organism: Marinomonas primoryensis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-09-01 Classification: PROTEIN BINDING Ligands: CA, 1PE |
 |
Organism: Marinomonas primoryensis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2021-09-01 Classification: PROTEIN BINDING Ligands: CA, 1PE |
 |
Fucose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: FUL, CA, EDO, FUC |
 |
Allose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, VDS, VDV |
 |
Mannose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, EDO, MAN, BMA |
 |
N-Acetyl-Glucosamine-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: NDG, NAG, CA, EDO |
 |
Inositol-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, EDO, INS |
 |
Sucrose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA |
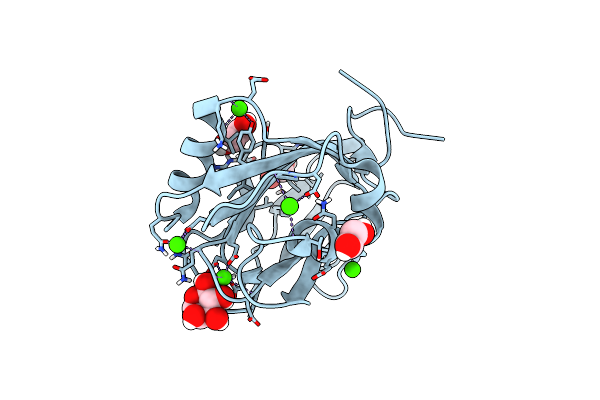 |
Arabinose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, ARA, EDO |
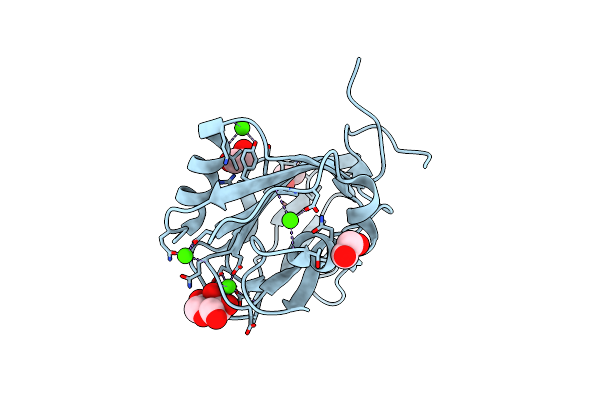 |
Ribose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: RIP, CA, EDO |
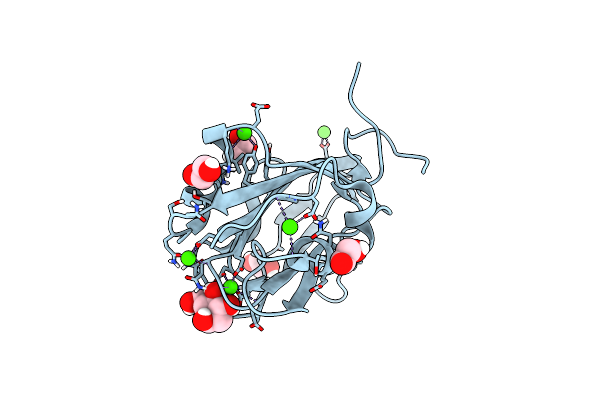 |
2-Deoxy-Glucose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: Z61, CA, EDO |
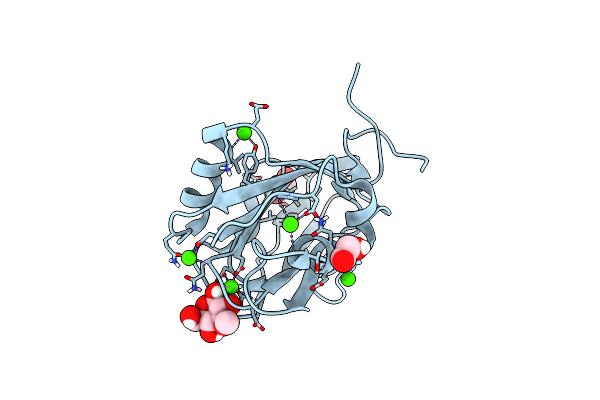 |
3-O-Methyl-Glucose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, EDO, 3MG |
 |
2-Deoxyribose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: UZJ, CA, EDO |

